| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Kalpana Mandal | + 2009 word(s) | 2009 | 2020-11-12 05:20:33 | | | |
| 2 | Vivi Li | -29 word(s) | 1980 | 2020-11-19 05:05:25 | | |
Video Upload Options
Phosphoinositides play a crucial role in regulating many cellular functions, such as actin dynamics, signaling, intracellular trafficking, membrane dynamics, and cell–matrix adhesion. Central to this process is phosphatidylinositol bisphosphate (PIP2). The levels of PIP2 in the membrane are rapidly altered by the activity of phosphoinositide-directed kinases and phosphatases, and it binds to dozens of different intracellular proteins. Despite the vast literature dedicated to understanding the regulation of PIP2 in cells over past 30 years, much remains to be learned about its cellular functions. Here, we focus on past and recent exciting results on different molecular mechanisms that regulate cellular functions by binding of specific proteins to PIP2 or by stabilizing phosphoinositide pools in different cellular compartments. Moreover, this review summarizes recent findings that implicate dysregulation of PIP2 in many diseases.
1. Introduction
Phosphoinositides (PPIs), are inositol-containing glycerophospholipids bearing variable numbers of phosphate groups on their headgroups [1]. PPIs are multifaceted molecules that have recently become an interesting player in regulating cell function due to their involvement in cellular functions such as actin dynamics, membrane trafficking, regulation of transmembrane proteins and signal transduction [2][3][4][5][6][7][8]. Although the total amount of PPIs in eukaryotic cell membranes is low, they play critical roles in cellular dynamics by regulating multiprotein complexes [9][10]. The spatiotemporal regulation of PPI-mediated biological processes is achieved by interconversion (Figure 1) of the phosphorylation states of PPIs by specific kinases and phosphatases, followed by recruitment of PPI-specific effectors. Inter-conversion of the phosphate groups is spatially controlled by phosphoinositide-metabolizing kinases and phosphatases as required for cellular functions [11][12]. PPIs generate seven possible isoforms by phosphorylating the inositol ring at positions 3, 4, and 5. Three isoforms of PPIs with two phosphate groups connected to the inositol ring, phosphatidylinositol-(4,5)-bisphosphate (PI(4, 5)P2), phosphatidylinositol-(3,5)-bisphosphate (PI(3, 5)P2) and phosphatidylinositol-(3,4)-bisphosphate (PI(3, 4)P2) are the focus of this review. PI(3,4)P2 and PI(3,5)P2 are produced by the phosphorylation of PI3P, and PI(4,5)P2 is produced by the phosphorylation of PI4P or PI5P. The synthesis of PIP2 by phosphorylation of PI5P is regulated by PIP4K, which is one of the less studied pathways. In addition, PIP4K is able to phosphorylate PI3P, but only able to alter PI5P levels in vivo [13] (PI(4, 5)P2) can be further phosphorylated to (PI(3,4,5)P3) and (PI(3,4,5)P3) is converted to (PI(4,5)P2) by the enzyme phosphate and tensin homolog deleted from chromosome 10 (PTEN) [14]. PIP3 stimulates the activity of PDK1 and phosphorylates Akt [15][16]. This PIP3/Akt pathway is intensively studied and regulates many crucial processes in cells. PIP3 and PTEN have been the subjects of excellent recent reviews, and the focus here is on PIP2 [7].
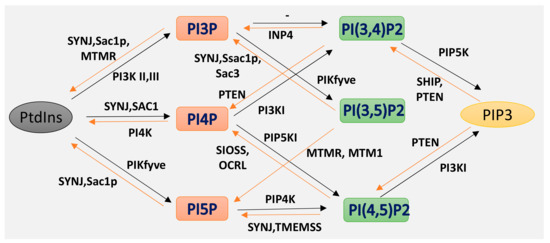
Figure 1. Isoforms of phosphoinositides. By the action of PIK and phosphatase, phosphatidylinositol (PtdIns) and the three isoforms of PIP2 are formed, as indicated here. The specific action of PI3K I, II III and of the 3-phosphatases are also illustrated.
Phosphoinositides control intracellular trafficking, membrane dynamics and cytoskeletal organization by interacting with many different proteins [17][18][19]. PIP2 regulates other membrane phospholipids and their signaling functions [7][17]. The major roles it plays in the cell membrane include cytoskeletal linkage, regulation of ion channels, and intracellular trafficking [20]. PI dynamics and mechanism are precisely controlled by kinase and phosphatase [21][22]. Recent studies showed the direct implication of these enzymes in diseases including liver cancer, glioblastoma or neurodegeneration [7][11][23]. Thus, many studies target phosphoinositide kinase inhibitors for pathological studies.
2. PIP2 in Actin Dynamics
Cytoskeletal dynamics play an important role in many cellular functions such as force generation, intracellular transport, or migration [24][25][26][27]. Actin forms the network inside the cell which is the most responsible for cellular architecture providing the cell a mechanical scaffold [24][27][28]. Accumulated evidence suggests that membrane PIP2 regulates the function of many acting binding proteins including formin, gelsolin, cofilin, profilin, filamin, WASP, ezrin, α- actinin, and others, which control the dynamical organization of the actin network [9][29][30][31][32][33]. PIP2 mostly inactivates actin binding proteins that inhibit actin polymerization and activates proteins which promote filamentous assembly [30][34]. Proteins bind to PIPs via numerus different structures, including the pleckstrin homology (PH) domain of phospholipase C-delta1, the Gag precursor protein Pr55 of HIV-1, phox homology (PX), C2, SH2, protein tyrosine binding, FYVE, PHD, GRAM, BAR, and espin N-terminal homology (ENTH)/ANTH domains, forming a large family of domains collectively [35][36].
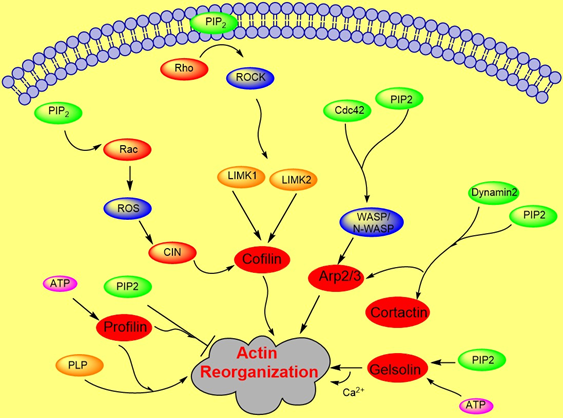
Actin polymerization is regulated by a variety of actin binding proteins [28][37]. Actin dynamics depend upon the continuous attachment of G- actin at the barbed (+) end and dissociation at the pointed (−) end, and that defines the filament length [38]. Cofilin is an actin binding protein that binds to both F-actin and G-actin and is a severing protein responsible for actin depolymerization (Figure 2) [38][39][40]. ADF/ cofilin binds to PIP2 through a multivalent mechanism and the dissociation of ADF/cofilin to actin filament can accurately be regulated by changing PIP2 density at the cell membrane [41]. One study found that cofilin binds to PIP2 via a specific pocket which is pH dependent [42]. However, this result contrasts with recent finding showing that cofilin interaction with PIP2 is not pH dependent, but the interaction of profilin with membrane, actin and multiple PIP2 headgroup (clustering) is affected somewhat when pH is increased [41]. Cofilin’s activity depends on phosphorylation, which is regulated by Rho-GTPase and LIM kinase (LIMK) and by binding PPIs [43]. The rho-family small GTPases, Rho, Rac, and Cdc42, play a central role in regulating actin reorganization through their various downstream effectors [44] LIMK1 and LIMK2 are activated by the GTPase-dependent protein kinases ROCK and PAK1 by phosphorylation of Thr508 and Thr-505, respectively, in the activation loop of the kinase domain [4][45]. LIMK1 and LIMK2 both regulate actin cytoskeletal reorganization by phosphorylating and inactivating cofilin/ADF [4][6]. Hence, cofilin is regulated by the signals from both the Rho and Rac pathways. Epidermal growth factor (EGF) induces sudden loss of PIP2 in membrane that activates local cofilin pool in membrane in carcinoma [46]. These altogether lead to a dramatic turnover of actin monomers (Figure 2).
Figure 2. Role of PIP2 in actin dynamics either by promoting polymerization or inhibiting severing. The figure summarizes gelsolin, profilin, cofilin, Arp2/3, and WASP dynamics in coordination with Rho- ROCK and Rac pathways.
3. PIP2 in Adhesion Dynamics
PIP2 binds to many focal adhesion proteins, such as vinculin, talin, and the focal adhesion kinase FAK. PIP2 serves as linkage to focal adhesion and actin binding proteins. There are actin binding proteins such as α-actinin, ezrin or filamin which also bind to focal adhesions. A synthetic peptide of α-actinin inhibits PLC-γ1 and PLC-δ1 activity and inhibition is induced by PIP2 competition [40]. PIP2 binding to α-actinin is inhibited by the treatment of cells with platelet derived growth factor, resulting in actin depolymerization. A recent study showed that the architecture of α-actinin-2 and 3 provides a suitable spatial orientation platform for PIP2 bonding by performing molecular dynamics (MD) simulations [47]. In smooth muscle in which α-actinin was discovered, PIP2 is found in large amounts which facilitates gelation of actin [48][49]. The length of smooth muscle depends upon the PIP2 synthesis, which regulates inositol phospholipid turnover [50]. Filamin A is another crosslinker protein which forms contacts between focal adhesions and F-actin. Filamin is associated to the cell membrane by β integrins. PIP2 bound to filamin A inhibits the gel formation of actin. Filamin has three isoforms called FLNa, FLNb, and FLNc. FLNa is recruited by CD28 followed by lipid raft accumulation at the immunological synapsis in T lymphocyte activation. PIP2 is essential for the clustering of lipid raft [51]. Ezrin is one of the ERM (ezrin, radixin, moesin) family proteins, which also forms linkages between the cellular membrane and cytoskeleton. Ezrin exists in both active and inactive states within cells. PIP2 activates Ezrin by binding with it and becomes available for phosphorylation by Rho-kinase and many PKC isoforms [52]. Neutron scattering experiment showed for the first-time, the conformational changes of ezrin when it simultaneously binds to PIP2 and F-actin [53].
Focal adhesion kinase (FAK) is a protein tyrosine kinase implicated in many signaling pathways to regulate cellular functions including migration. When a cell binds to the extracellular matrix (ECM), FAK is recruited to focal adhesion (FA) sites and undergoes conformational change, which is activated by phospholipids such as PIP2 by unblocking the FERM domain and kinase domain. Simulation results show that FAK transiently binds to PIP2 through electrostatic interactions [54]. Molecular dynamics simulation and fluorescence resonance energy transfer (FRET) experiments both showed that FAK binding to ATP decreases the FRET signal confirming that the PIP2 binding acts in the reverse direction [55][56]. Phosphatidylinositol 4-phosphate 5-kinase type Iγ (PIP5KIγ) is required for efficient FAK activation and generates PIP2 locally in FAs by PIP5KIγ. Thus, PIP2 is a strong mediator in integrin-FAK signaling pathways [55].
Talin plays a crucial role in activating integrins [57][58]. Within the cytosol talin is in an inactivated form, where its C-terminal rod domain binds to the N-terminal head domain. Many pathways lead to disruption of the interaction between talin’s C-terminal and N-terminal including binding with PIP5KIγ which generates PIP2 from PI4P [59]. Ye et al. delineate a detailed account of PIP2 in activating talin by using FRET. They showed interaction of talin with lipid bilayers is altered by PIP2 [60]. The FERM domain of talin-1 binds to the cytosolic domain of β3 -integrin weakly (Figure 3). However, the interaction affinity increases three-fold when it synergistically binds to acidic PIP2 [8][61][62]. Membrane bound talin recruits and activates vinculin. Vinculin localizes at the adhesion complex and interacts with PIP2 to associate with the membrane (Figure 3) [63]. Simulation data shows that PIP2 is not required for vinculin localization at FAs but is needed for the activation of FA turnover during mechanotransduction processes [63]. Other studies mentioned that PIP2 is required for FA formation and vinculin phosphorylation and trafficking [64].
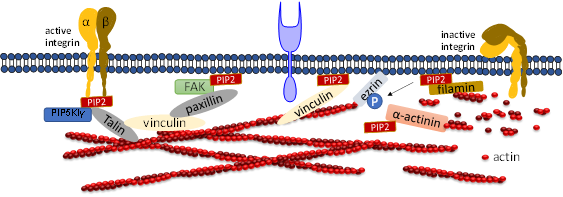
Figure 3. Role of PIP2 in regulating focal adhesion assembly. Depiction of adhesion molecules talin, vinculin, ezrin, filamin and a-actinin. PIP2 synergistically binds to both talin and integrin and activates both of them. Talin binds directly to actin or activates vinculin and facilitates its binding to actin. PIP2 also binds to FERM domain of FAK and binds to vinculin via paxillin. PIP2 negatively regulates cross-linking activity of filamin and the actin bundle formation mediated by α-actinin.
4. Conclusions and outlook
Past evidence suggests that actin is connected to the membrane via actin binding proteins such as α-actinin or filamin which are regulated by phosphoinositides. These interactions also affect the binding of actin filaments with focal adhesion proteins such as paxillin, talin, FAK, or vinculin. The distribution of PIP2 in the membrane regulates cell signaling. PIP2 activity depends upon the concentration of cholesterol and divalent ions such as Ca2+, Mg2+, or Zn2+. In addition, PIP2 plays a crucial role in modulating many signaling pathways such as PIP3/Akt, mTORc1, or Rho dependent pathways that have implications in many diseases including cancer, neurodegenerative disease, or down syndrome.
Although PPIs are essential for many cellular functions, there are disparities in many processes which need further studies. PIP2 plays an important role in actin reorganization and filament dynamics. However, the role of PIP2 in any other cytoskeletal component has not yet been well studied. Among PIP2 binding actin proteins, LIMK1 and LIMK2 play an overlapping role in actin reorganization in the Rho-ROCK pathway. Further studies are required to differentiate the functional role of LIMK1 and LIMK2. Moreover, it is unclear if members of ROCK and PAK family proteins function as LIMK- activating kinases. Cortactin shows dependencies on PIP2 and Rac in dissociating from actin–myosin complex, although the direct implication of PIP2 in regulating cortactin still remains controversial [65], and other activators such as the endocytic protein Abp1p remain unclear. It has been shown that a synthetic peptide of α-actinin inhibits PLC-γ1 and PLC-δ1. It is ambiguous whether PIP2 bound to α-actinin is hydrolyzed by activated PLC-γ1 or not. The interaction of vinculin and membrane is based upon either full length or tail domain of vinculin in lipid bilayers or in cells. However, a specific lipid binding site has yet to be discovered.
References
- Downes, C.P.; Gray, A.; Lucocq, J.M. Probing phosphoinositide functions in signaling and membrane trafficking. Trends Cell Biol. 2005, 15, 259–268.
- Picas, L.; Viaud, J.; Schauer, K.; Vanni, S.; Hnia, K.; Fraisier, V.; Roux, A.; Bassereau, P.; Gaits-Iacovoni, F.; Payrastre, B.; et al. BIN1/M-Amphiphysin2 induces clustering of phosphoinositides to recruit its downstream partner dynamin. Nat. Commun. 2014, 5, 1–12.
- McLean, M.A.; Stephen, A.G.; Sligar, S.G. PIP2 Influences the Conformational Dynamics of Membrane-Bound KRAS4b. Biochemistry 2019, 58, 3537–3545.
- Romarowski, A.; Battistone, M.A.; La Spina, F.A.; del Puga Molina, L.C.; Luque, G.M.; Vitale, A.M.; Cuasnicu, P.S.; Visconti, P.E.; Krapf, D.; Buffone, M.G. PKA-dependent phosphorylation of LIMK1 and Cofilin is essential for mouse sperm acrosomal exocytosis. Dev. Biol. 2015, 405, 237–249.
- Liepiņa, I.; Czaplewski, C.; Janmey, P.; Liwo, A. Molecular dynamics study of a gelsolin-derived peptide binding to a lipid bilayer containing phosphatidylinositol 4,5-bisphosphate. Biopolymers 2003, 71, 49–70.
- Amano, T.; Tanabe, K.; Eto, T.; Narumiya, S.; Mizuno, K. LIM-kinase 2 induces formation of stress fibres, focal adhesions and membrane blebs, dependent on its activation by Rho-associated kinase-catalysed phosphorylation at threonine-505. Biochem. J. 2001, 354, 149–159.
- Walter, L.M.; Franz, P.; Lindner, R.; Tsiavaliaris, G.; Hensel, N.; Claus, P. Profilin2a-phosphorylation as a regulatory mechanism for actin dynamics. FASEB J. 2020, 34, 2147–2160.
- Saltel, F.; Mortier, E.; Hytönen, V.P.; Jacquier, M.C.; Zimmermann, P.; Vogel, V.; Liu, W.; Wehrle-Haller, B. New PI(4,5)P2- and membrane proximal integrin-binding motifs in the talin head control β3-integrin clustering. J. Cell Biol. 2009, 187, 715–731.
- Janmey, P.A.; Bucki, R.; Radhakrishnan, R. Regulation of actin assembly by PI(4,5)P2 and other inositol phospholipids: An update on possible mechanisms. Biochem. Biophys. Res. Commun. 2018, 506, 307–314.
- Reversi, A.; Loeser, E.; Subramanian, D.; Schultz, C.; De Renzis, S. Plasma membrane phosphoinositide balance regulates cell shape during Drosophila embryo morphogenesis. J. Cell Biol. 2014, 205, 395–408.
- Viaud, J.; Mansour, R.; Antkowiak, A.; Mujalli, A.; Valet, C.; Chicanne, G.; Xuereb, J.M.; Terrisse, A.D.; Séverin, S.; Gratacap, M.P.; et al. Phosphoinositides: Important lipids in the coordination of cell dynamics. Biochimie 2016, 125, 250–258.
- Goud, B.; Picas, L.; Gaits-Iacovoni, F. The emerging role of phosphoinositide clustering in intracellular trafficking and signal transduction. F1000Research 2016, 5.
- Rameh, L.E.; Tolias, K.F.; Duckworth, B.C.; Cantley, L.C. A new pathway for synthesis of phosphatidylinositol-4,5-bisphosphate. Nature 1997, 390, 192–196.
- Liu, C.; Deb, S.; Ferreira, V.S.; Xu, E.; Baumgart, T. Kinetics of PTEN-mediated PI(3,4,5)P3 hydrolysis on solid supported membranes. PLoS ONE 2018, 13, e0192667.
- Thillai, K.; Lam, H.; Sarker, D.; Wells, C.M. Deciphering the link between PI3K and PAK: An opportunity to target key pathways in pancreatic cancer? Oncotarget 2017, 8, 14173–14191.
- Vasudevan, K.M.; Barbie, D.A.; Davies, M.A.; Rabinovsky, R.; McNear, C.J.; Kim, J.J.; Hennessy, B.T.; Tseng, H.; Pochanard, P.; Kim, S.Y.; et al. AKT-independent signaling downstream of oncogenic PIK3CA mutations in human cancer. Cancer Cell 2009, 16, 21–32.
- Balla, T. Phosphoinositides: Tiny lipids with giant impact on cell regulation. Physiol. Rev. 2013, 93, 1019–1137.
- Goud, B.; Liu, S.; Storrie, B. Rab proteins as major determinants of the Golgi complex structure. Small GTPases 2018, 9, 66–75.
- Heinrich, M.; Tian, A.; Esposito, C.; Baumgart, T. Dynamic sorting of lipids and proteins in membrane tubes with a moving phase boundary. Proc. Natl. Acad. Sci. USA 2010, 107, 7208–7213.
- Huang, C.L. Complex roles of PIP2 in the regulation of ion channels and transporters. Am. J. Physiol. Ren. Physiol. 2007, 293, 1761–1765.
- Rodriguez-Menchaca, A.A.; Adney, S.K.; Tang, Q.-Y.; Meng, X.-Y.; Rosenhouse-Dantsker, A.; Cui, M.; Logothetis, D.E.; Designed, D.E.L.; Performed, M.C. PIP 2 controls voltage-sensor movement and pore opening of Kv channels through the S4-S5 linker. Proc. Natl. Acad. Sci. USA 2012, 109, E2399–E2408.
- Zaydman, M.A.; Silva, J.R.; Delaloye, K.; Li, Y.; Liang, H.; Larsson, H.P.; Shi, J.; Cui, J. Kv7.1 ion channels require a lipid to couple voltage sensing to pore opening. Proc. Natl. Acad. Sci. USA 2013, 110, 13180–13185.
- Erle, D.J.; Sheppard, D. The cell biology of asthma. J. Cell Biol. 2014, 205, 621–631.
- Mandal, K.; Wang, I.; Vitiello, E.; Orellana, L.A.C.; Balland, M. Cell dipole behaviour revealed by ECM sub-cellular geometry. Nat. Commun. 2014, 5.
- Guet, D.; Mandal, K.; Pinot, M.; Hoffmann, J.; Abidine, Y.; Sigaut, W.; Bardin, S.; Schauer, K.; Goud, B.; Manneville, J.B. Mechanical role of actin dynamics in the rheology of the Golgi complex and in Golgi-associated trafficking events. Curr. Biol. 2014, 24, 1700–1711.
- Mizuno, D.; Tardin, C.; Schmidt, C.F.; Mackintosh, F.C. Nonequilibrium mechanics of active cytoskeletal networks. Science 2007, 315, 370–373.
- Mandal, K.; Balland, M.; Bureau, L. Thermoresponsive micropatterned substrates for single cell studies. PLoS ONE 2012, 7, e37548.
- Mandal, K.; Asnacios, A.; Goud, B.; Manneville, J.-B. Mapping intracellular mechanics on micropatterned substrates. Proc. Natl. Acad. Sci. USA 2016, 113, E7159–E7168.
- Honigmann, A.; Van Den Bogaart, G.; Iraheta, E.; Risselada, H.J.; Milovanovic, D.; Mueller, V.; Müllar, S.; Diederichsen, U.; Fasshauer, D.; Grubmüller, H.; et al. Phosphatidylinositol 4,5-bisphosphate clusters act as molecular beacons for vesicle recruitment. Nat. Struct. Mol. Biol. 2013, 20, 679–686.
- Yin, H.L.; Janmey, P.A. Phosphoinositide Regulation of the Actin Cytoskeleton. Annu. Rev. Physiol. 2003, 65, 761–789.
- Pogoda, K.; Bucki, R.; Byfield, F.J.; Cruz, K.; Lee, T.; Marcinkiewicz, C.; Janmey, P.A. Soft Substrates Containing Hyaluronan Mimic the Effects of Increased Stiffness on Morphology, Motility, and Proliferation of Glioma Cells. Biomacromolecules 2017, 18, 3040–3051.
- Gowrishankar, K.; Ghosh, S.; Saha, S.; Mayor, S.; Rao, M. Active Remodeling of Cortical Actin Regulates Spatiotemporal Organization of Cell Surface Molecules. Cell 2012, 149, 1353–1367.
- Mandal, K.; Raz-Ben Aroush, D.; Graber, Z.T.; Wu, B.; Park, C.Y.; Fredberg, J.J.; Guo, W.; Baumgart, T.; Janmey, P.A. Soft Hyaluronic Gels Promote Cell Spreading, Stress Fibers, Focal Adhesion, and Membrane Tension by Phosphoinositide Signaling, Not Traction Force. ACS Nano 2018, acsnano.8b05286.
- Yamamoto, M.; Hilgemann, D.H.; Feng, S.; Bito, H.; Ishihara, H.; Shibasaki, Y.; Yin, H.L. Phosphatidylinositol 4,5-bisphosphate induces actin stress-fiber formation and inhibits membrane ruffling in CV1 cells. J. Cell Biol. 2001, 152, 867–876.
- Lemmon, M.A. Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol. 2008, 9, 99–111.
- Levine, T.P.; Munro, S. The pleckstrin homology domain of oxysterol-binding protein recognises a determinant specific to Golgi membranes. Curr. Biol. 1998, 8, 729–739.
- Senju, Y.; Kalimeri, M.; Koskela, E.V.; Somerharju, P.; Zhao, H.; Vattulainen, I.; Lappalainen, P. Mechanistic principles underlying regulation of the actin cytoskeleton by phosphoinositides. Proc. Natl. Acad. Sci. USA 2017, 114, 8977–8986.
- Bugyi, B.; Carlier, M.-F. Control of Actin Filament Treadmilling in Cell Motility. Ann. Rev. Biophys. 2010, 39, 449–470.
- Bernstein, B.W.; Bamburg, J.R. ADF/Cofilin: A functional node in cell biology. Trends Cell Biol. 2010, 20, 187–195.
- Sakisaka, T.; Itoh, T.; Miura, K.; Takenawa, T. Phosphatidylinositol 4,5-bisphosphate phosphatase regulates the rearrangement of actin filaments. Mol. Cell. Biol. 1997, 17, 3841–3849.
- Zhao, H.; Hakala, M.; Lappalainen, P. ADF/cofilin binds phosphoinositides in a multivalent manner to act as a PIP(2)-density sensor. Biophys. J. 2010, 98, 2327–2336.
- Leyman, S.; Sidani, M.; Ritsma, L.; Waterschoot, D.; Eddy, R.; Dewitte, D.; Debeir, O.; Decaestecker, C.; Vandekerckhove, J.; Van Rheenen, J.; et al. Unbalancing the phosphatidylinositol-4,5-bisphosphate-cofilin interaction impairs cell steering. Mol. Biol. Cell 2009, 20, 4509–4523.
- Sumi, T.; Matsumoto, K.; Takai, Y.; Nakamura, T. Cofilin phosphorylation and actin cytoskeletal dynamics regulated by Rho- and Cdc42-activated LIM-kinase 2. J. Cell Biol. 1999, 1519–1532.
- Croisé, P.; Eestay-Aahumada, C.; Gasman, S.; Ory, S. Rho GTPases, phosphoinositides, and actin: A tripartite framework for efficient vesicular trafficking. Small GTPases 2014, 5.
- Prunier, C.; Prudent, R.; Kapur, R.; Sadoul, K.; Lafanechère, L. LIM kinases: Cofilin and beyond. Oncotarget 2017, 8, 41749–41763.
- Van Rheenen, J.; Song, X.; Van Roosmalen, W.; Cammer, M.; Chen, X.; DesMarais, V.; Yip, S.C.; Backer, J.M.; Eddy, R.J.; Condeelis, J.S. EGF-induced PIP2 hydrolysis releases and activates cofilin locally in carcinoma cells. J. Cell Biol. 2007, 179, 1247–1259.
- Ribeiro, E.D.A.; Pinotsis, N.; Ghisleni, A.; Salmazo, A.; Konarev, P.V.; Kostan, J.; Sjöblom, B.; Schreiner, C.; Polyansky, A.A.; Gkougkoulia, E.A.; et al. The structure and regulation of human muscle α-Actinin. Cell 2014, 159, 1447–1460.
- Pavalko, F.M.; Otey, C.A.; Simon, K.O.; Burridge, K. α-Actinin: A direct link between actin and integrins. Biochem. Soc. Trans. 1991, 19, 1065–1069.
- Fukami, K.; Sawada, N.; Endo, T.; Takenawa, T. Identification of a Phosphatidylinositol 4,5-Bisphosphate-binding Site in Chicken Skeletal Muscle α-Actinin. J. Biol. Chem. 1996, 271, 2646–2650.
- Sul, D.; Baron, C.B.; Broome, R.; Coburn, R.F. Smooth muscle length-dependent PI(4,5)P2 synthesis and paxillin tyrosine phosphorylation. Am. J. Physiol. Cell Physiol. 2001, 281, 300–310.
- Kallikourdis, M.; Trovato, A.E.; Roselli, G.; Muscolini, M.; Porciello, N.; Tuosto, L.; Viola, A. Phosphatidylinositol 4-Phosphate 5-Kinase β Controls Recruitment of Lipid Rafts into the Immunological Synapse. J. Immunol. 2016, 196, 1955–1963.
- Shabardina, V.; Kramer, C.; Gerdes, B.; Braunger, J.; Cordes, A.; Schäfer, J.; Mey, I.; Grill, D.; Gerke, V.; Steinem, C. Mode of Ezrin-Membrane Interaction as a Function of PIP2 Binding and Pseudophosphorylation. Biophys. J. 2016, 110, 2710–2719.
- Jayasundar, J.J.; Ju, J.H.; He, L.; Liu, D.; Meilleur, F.; Zhao, J.; Callaway, D.J.E.; Bu, Z. Open conformation of ezrin bound to phosphatidylinositol 4,5-bisphosphate and to F-actin revealed by neutron scattering. J. Biol. Chem. 2012, 287, 37119–37133.
- Feng, J.; Mertz, B. Novel Phosphotidylinositol 4,5-Bisphosphate Binding Sites on Focal Adhesion Kinase. PLoS ONE 2015, 10, e0132833.
- Goñi, G.M.; Epifano, C.; Boskovic, J.; Camacho-Artacho, M.; Zhou, J.; Bronowska, A.; Martín, M.T.; Eck, M.J.; Kremer, L.; Gräter, F.; et al. Phosphatidylinositol 4,5-bisphosphate triggers activation of focal adhesion kinase by inducing clustering and conformational changes. Proc. Natl. Acad. Sci. USA 2014, 111, E3177–E3186.
- Zhou, J.; Bronowska, A.; Le Coq, J.; Lietha, D.; Gräter, F. Allosteric regulation of focal adhesion kinase by PIP2 and ATP. Biophys. J. 2015, 108, 698–705.
- Mcnamee, H.P.; Liley, H.G.; Ingber, D.E. Integrin-Dependent Control of Inositol Lipid Synthesis in Vascular Endothelial Cells and Smooth Muscle Cells. Exp. Cell Res. 1996, 224, 116–122.
- Borowsky, M.L.; Hynes, R.O. Layilin, a novel talin-binding transmembrane protein homologous with C-type lectins, is localized in membrane ruffles. J. Cell Biol. 1998, 143, 429–442.
- Wang, Y.; Zhao, L.; Suzuki, A.; Lian, L.; Min, S.H.; Wang, Z.; Litvinov, R.I.; Stalker, T.J.; Yago, T.; Klopocki, A.G.; et al. Platelets lacking PIP5KIγ have normal integrin activation but impaired cytoskeletal-membrane integrity and adhesion. Blood 2013, 121, 2743–2752.
- Das, M.; Ithychanda, S.; Qin, J.; Plow, E.F. Mechanisms of talin-dependent integrin signaling and crosstalk. Biochim. Biophys. Acta 2014, 1838, 579–588.
- Moore, D.T.; Nygren, P.; Jo, H.; Boesze-Battaglia, K.; Bennett, J.S.; DeGrado, W.F. Affinity of talin-1 for the β3-integrin cytosolic domain is modulated by its phospholipid bilayer environment. Proc. Natl. Acad. Sci. USA 2012, 109, 793–798.
- Elliott, P.R.; Goult, B.T.; Kopp, P.M.; Bate, N.; Grossmann, J.G.; Roberts, G.C.K.; Critchley, D.R.; Barsukov, I.L. The Structure of the Talin Head Reveals a Novel Extended Conformation of the FERM Domain. Structure 2010, 18, 1289–1299.
- Thompson, P.M.; Ramachandran, S.; Case, L.B.; Tolbert, C.E.; Tandon, A.; Pershad, M.; Dokholyan, N.V.; Waterman, C.M.; Campbell, S.L. A Structural Model for Vinculin Insertion into PIP2-Containing Membranes and the Effect of Insertion on Vinculin Activation and Localization. Structure 2017, 25, 264–275.
- Chandrasekar, I.; Stradal, T.E.B.; Holt, M.R.; Entschladen, F.; Jockusch, B.M.; Ziegler, W.H. Vinculin acts as a sensor in lipid regulation of adhesion-site turnover. J. Cell Sci. 2005, 118, 1461–1472.
- Daly, R.J. Cortactin signalling and dynamic actin networks. Biochem. J. 2004, 382, 13–25.