| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Sourya Subhra Nasker | + 1757 word(s) | 1757 | 2020-12-28 03:55:07 | | | |
| 2 | Bruce Ren | -21 word(s) | 1736 | 2021-01-26 02:11:15 | | |
Video Upload Options
Inteins are mobile genetic elements that apply standard enzymatic strategies to excise themselves post-translationally from the precursor protein via protein splicing. Since their discovery in the 1990s, recent advances in intein technology allow for them to be implemented as a modern biotechnological contrivance. Radical improvement in the structure and catalytic framework of cis- and trans-splicing inteins devised the development of engineered inteins that contribute to various efficient downstream techniques.
1. Introduction
Splicing mechanism can be broadly categorized as RNA splicing and protein splicing, two mechanisms responsible for the flow of information from a gene to its protein product to yield a functional protein whose sequence is strictly noncolinear with the gene. While group I introns self-splice at a precursor RNA level, intein splicing involves the removal of an intervening sequence at a precursor polypeptide level [1]. Initially, this intervening polypeptide sequence was termed as spacer or protein introns, currently termed as inteins (INTervening protEINS) [2]. In a radical post-translational event, inteins excise themselves precisely from a larger precursor protein by sequential cleavage of peptide bonds and concomitant ligation by peptide bond formation between the flanking amino-terminal (N-) and carboxy-terminal (C-) residues termed as exteins, resulting in the formation of an active protein product [3][4]. The intein-mediated splicing mechanism lacks the use of any exogenous cofactors or high-energy molecules [5]. The embodiment of intein-mediated protein splicing in the “central dogma” of molecular biology puts in an additional level of complexity to the mechanism of gene expression [6].
Most of the inteins are interrupted by a homing endonuclease domain (HED) [7][8][9]. However, the HED can be removed from within the intein, without entirely compromising the splicing activity [10]. Thus the presence of HEDs increases the allele frequency at a rate higher than that of Mendelian rates [11]. Homing endonucleases encoded within an intein contains the prefix “PI-” in terms of intein nomenclature [12][13]. Conventionally, intein nomenclature comprises abbreviated names of both genus and species followed by the name of the protein; an intein residing in the GyrA protein of Mycobacterium xenopi is designated as Mxe GyrA, for instance [14]. Mxe GyrA, coincidentally, is also the smallest known naturally occurring intein [15].
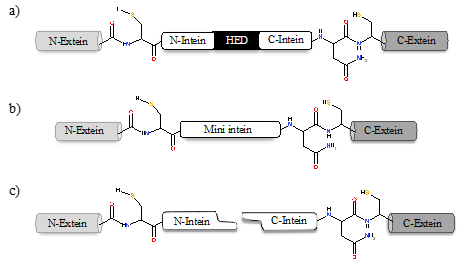
Inteins naturally exist in three different configurations (Figure 1): (1) full-length inteins, where a sequence-specific homing endonuclease domain is embedded in between the splicing (catalytic) domains; (2) mini-inteins, lacking the homing endonuclease domain and containing a contiguous protein splicing domain; and (3) split inteins, transcribed and translated as two separate polypeptides each joined with an extein [16][17]. The study of intein distribution, dissemination, and their potential biological functions are particularly fascinating in the field of translational research. Inteins distribution is sporadic in the genomes of organisms spanning from archaea, bacteria and eukaryotes to several viral genomes [18][19][20]. The reason for such anomalous distribution has spurred the proposal for numerous evolutionary scenarios, including the role of inteins in genetic mobility and as a selfish DNA [19][20][21]. Still, the question remains as to why inteins persisted for millions of years? Do they perform a beneficial role in the host or are they just a selfish gene? This phenomenon is puzzling and needs to be explored further.
Figure 1. Intein configurations. Schematic representation of various types of intein: (a) full-length intein with Homing endonuclease domain (HED), (b) mini-intein, and (c) split intein.
The potential to exploit inteins for a practical purpose has led to the development of a diverse array of applications in modern biotechnology. Inteins can be engineered to undergo conditional protein splicing (CPS) which requires environmental or molecular triggers like light, changes in pH or temperature, change in redox state, or addition of small molecules [22][23][24][25][26][27]. The bias nature of inteins toward plant and human pathogens makes it an attractive tool for novel drug development [28][29][30]. The development of engineered inteins or synthetic intein systems has encouraged efficient protein purification, ligation, and cyclization strategies [31][32]. Recent advances in intein research have extended these in vitro application to whole organisms. Such developing applications suggest that inteins are becoming a mature and critical biological tool, capable of widening the aperture to new avenues of scientific research, including enhanced transgenic plants and novel therapeutic strategies [33].
2. Intein Distribution and Evolution
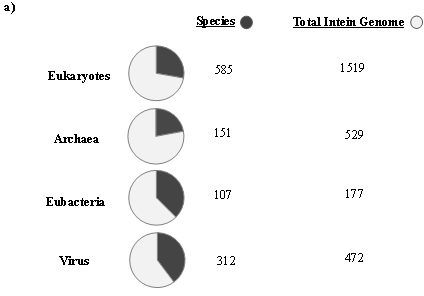
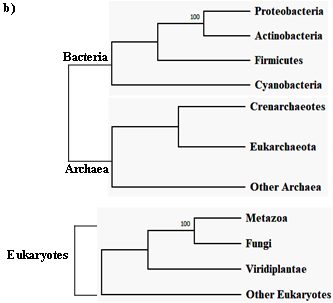
The first intein sequence was discovered 32 years ago in the Saccharomyces cerevisiae VMA1 gene that encodes for an alpha subunit of vacuolar H+ ATPase [34][35]. The translational product of the gene was calculated to be 118.6 KDa but experimentally estimated as 67 KDa. The deduced amino acid sequence shows similarity to other ATPase at N- and C- terminal regions, but the central region was not determined [34]. Experimental analysis by Kane et al. revealed the presence of two separate proteins of molecular weights 69 and 50 KDa [35]. Since then, further examples of inteins were found in all three domains of life—in archaea, the DNA polymerase of the extremely thermophilic archaebacteria Thermococcus litoralis [36], in bacteria, the RecA proteins of M. tuberculosis [37][38] and M. leprae [39] and in eukarya, the 69 KDa subunit of vacuolar ATPase of the yeast Candida tropicalis [40]. This highlights a wider distribution of inteins across all three domains of life (Figure 2b), suggesting an ancient origin that predates the separation of prokaryotes and eukaryotes [41]. We dug into the NCBI Gene database (www.ncbi.nlm.nih.gov/gene) to scan the distribution of intein in all the three domains of life, where out of 2709 intein-containing genomes, 56% of the total intein-containing genome is found in eukaryotes, 19.8% in archaea, and 6.64% in eubacteria. We also performed an assessment for intein distribution in viruses and observed 17.4% of the total intein-containing genome is present in viruses (Figure 2a).
Figure 2. Sporadic distribution of inteins. (a) Summary of intein distribution with the total number of intein-containing genome from respective species indicated. The intein distribution data were extracted from the NCBI Gene database. (b) Schematic representation of the tree of life showing four phyla for bacteria, three phyla for archaea, and three kingdoms of eukarya (Metazoa, Fungi, and Viridiplantae). All other eukaryotes are shown with the basal branch. Intein-containing gene sequences were obtained from NCBI and analyzed by MEGA-X software. The phylogenetic tree was constructed using the neighbor-joining method.
Novikova et al. performed a large-scale survey in order to analyze intein presence across bacteria and archaea. The survey revealed that half of the total archaeal genomes analyzed had at least one intein; in contrast, only a quarter of bacteria were found to be intein positive among the total bacterial genome studied. A recent study conducted by Kelly et al. sheds light on intein distribution across bacteria and their phages. This analysis provides the first clear evidence of mycobacteriophages as major facilitators of intein dissemination across all of mycobacteria. The study found that 19.1% of mycobacteriophages contain inteins residing mostly in nucleic acid binding proteins, enriched in specific clusters [42]. Regardless of the exiguous presence of inteins in eukaryotes as reported by bioinformatics analysis, there is, however, intein presence in the fungal nuclear genome, algal chloroplast genome and within few eukaryotic viruses. There is, however, a preponderance of inteins observed in fungi, mostly in Ascomycota representing some noteworthy pathogenic fungi, such as Candida sp. and Aspergillus sp. Among others, inteins found in Basidiomycota include human pathogens, such as Cryptococcus neoformans and C. gattii [43][44][45] and plant pathogens Tilletia indica and T. walkeri [46]. The chloroplast DNA of diverse algae and seaweeds contains a staggering number of inteins in the Rhodophyta, Chlorophyta, Cryptophyta, Ochrophyta and Heterokonta phylums . Amidst known eukaryotic viruses, there are hundreds of intein across four families, namely, Iridoviridae, Marseilleviridae, Phycodnaviridae and Mimiviridae [47]. Aforementioned fungal pathogens have intein presence commonly in Prp8 (pre-mRNA processing factor 8), VMA1 (vacuolar ATPase, subunit A), DnaB (DNA replication helicase DnaB-like), DdRP (RNA polymerase subunit beta RpoB), DdDP (DNA polymerases) and RIR (Ribonucleoside-diphosphate reductases).
The primary indication of intein origin lies in its two-domain structure, suggesting that a mobile intein is a result of a fusion between two proteins, most likely, a self-splicing intein and an endonuclease protein. Sequence and mutational studies reported that the endonuclease activity is concentrated in the central portion of the intein, whereas the splicing activity is located in the two-terminal regions [48][49]. However, it remains unclear whether an intein came first or the autocatalytic self-splicing domain in regulatory proteins. Xiang-Qin Liu stated that a self-splicing mini-intein shows a correspondence between its structural and functional composition. A mini-intein structurally consists of two subdomains along with a loop exchange between the same. Functionally, the splicing pathway consists of two peptide cleavages and a coupling between the two cleavages. This is not rather coincidental but suggests a structure-function relationship of the mini-intein. Liu further hypothesized that a fusion between two coding sequences gives rise to a duplication event in the domain responsible for the self-cleaving activity. This fusion protein retains its biological property to perform self-cleavage independently. It may be that the homing endonucleases invade such an element later on. This idea is supported by the reason that endonucleases, being mobile in the genome, although remove themselves from the gene product but would account for a preferable integration site in these locations since the function encoded by the surrounding genetic elements would not be disrupted. It is reasonable to think that naturally occurring mini-inteins most likely evolved from bifunctional mobile inteins by losing their endonuclease domain because once an intein enters a host protein, there is no considerable selection pressure to maintain endonuclease activity, but a strong selection pressure for maintaining the splicing activity. A split-intein may evolve from a mini-intein by initiating a break in the intein’s coding region. The discovery of naturally occurring split-intein in a cyanobacterial DNA polymerase (DnaE) supports the idea. The N- and C-exteins of DnaE are linked to their respective intein fragment. It is, however, encoded by two separate genes located on different parts of the genome [50].
Interestingly, inteins are biased towards invading regulatory proteins that are responsible for DNA metabolisms (polymerases, topoisomerases, helicases, ribonucleotide reductases) and essential housekeeping genes, including essential proteases, metabolic enzymes, RNA processing proteins, and energy supplying vital proteins. Their insertion site coincides with the conserved domains, responsible for host protein function like catalytic or ligand binding sites, enzyme active site, DNA binding sites etc. Insertion at these critical sites ensures the survivability of inteins, making them less prone to deletions. This site-specific behavior of intein insertion may be due to the functionality of its homing endonuclease domain. The amount of information conceived regarding the genome organization and expression of inteins in the last two decades has led to the understanding as to how mobile genetic elements are not solely parasitic sequences, but also have a dynamic role in the evolution of species.
References
- Derbyshire: V.; Belfort, M. Lightning strikes twice: Intron–intein coincidence. Proc. Natl. Acad. Sci. USA 1998, 95, 1356–1357.
- Gogarten, J.P.; Senejani, A.G.; Zhaxybayeva, O.; Olendzenski, L.; Hilario, E. Inteins: Structure, function, and evolution. Annu. Rev. Microbiol. 2002, 56, 263–287.
- Mills, K.V.; Johnson, M.A.; Perler, F.B. Protein splicing: How inteins escape from precursor proteins. J. Biol. Chem. 2014, 289, 14498–14505.
- Shah, N.H.; Muir, T.W. Inteins: nature’s gift to protein chemists. Chem. Sci. 2014, 5, 446–461.
- Paulus, H. Inteins as enzyme. Bioorganic Chem. 2001, 29, 119–129.
- Shao, Y.; Kent, S.B. Protein splicing: Occurrence, mechanisms and related phenomena. Chem. Biol. 1997, 4, 187–194.
- Barzel, A.; Naor, A.; Privman, E.; Kupiec, M.; Gophna, U. Homing endonucleases residing within inteins: Evolutionary puzzles awaiting genetic solutions. Biochem. Soc. Trans. 2011, 39, 169–173.
- Elleuche, S.; Pöggeler, S. Inteins, valuable genetic elements in molecular biology and biotechnology. Appl. Microbiol. Biotechnol. 2010, 87, 479–489.
- Topilina, N.I.; Mills, K.V. Recent advances in in vivo applications of intein-mediated protein splicing. Mob. Dna 2014, 5, 1–14.
- Iwaï, H.; Mikula, K.M.; Oeemig, J.S.; Zhou, D.; Li, M.; Wlodawer, A. Structural basis for the persistence of homing endonucleases in transcription factor IIB inteins. J. Mol. Biol. 2017, 429, 3942–3956.
- Yahara, K.; Fukuyo, M.; Sasaki, A.; Kobayashi, I. Evolutionary maintenance of selfish homing endonuclease genes in the absence of horizontal transfer. Proc. Natl. Acad. Sci. USA 2009, 106, 18861–18866.
- Belfort, M.; Roberts, R.J. Homing endonucleases: Keeping the house in order. Nucleic Acids Res. 1997, 25, 3379–3388.
- Perler, F.B. InBase: The intein database. Nucleic Acids Res. 2002, 30, 383–384.
- Perler, F.B.; Davis, E.O.; Dean, G.E.; Gimble, F.S.; Jack, W.E.; Neff, N.; Noren, C.J.; Thorner, J.; Belfort, M. Protein splicing elements: Inteins and exteins—A definition of terms and recommended nomenclature. Nucleic Acids Res. 1994, 22, 1125.
- Telenti, A.; Southworth, M.; Alcaide, F.; Daugelat, S.; Jacobs, W.R.; Perler, F.B. The Mycobacterium xenopi GyrA protein splicing element: Characterization of a minimal intein. J. Bacteriol. 1997, 179, 6378–6382.
- Fernandes, J.A.; Prandini, T.H.; Castro, M.d.C.A.; Arantes, T.D.; Giacobino, J.; Bagagli, E.; Theodoro, R.C. Evolution and application of inteins in Candida species: A review. Front. Microbiol. 2016, 7, 1585.
- Belfort, M.; Stoddard, B.L.; Wood, D.W.; Derbyshire, V. Homing Endonucleases and Inteins; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2006; Volume 16.
- Novikova, O.; Jayachandran, P.; Kelley, D.S.; Morton, Z.; Merwin, S.; Topilina, N.I.; Belfort, M. Intein clustering suggests functional importance in different domains of life. Mol. Biol. Evol. 2016, 33, 783–799.
- Novikova, O.; Topilina, N.; Belfort, M. Enigmatic distribution, evolution, and function of inteins. J. Biol. Chem. 2014, 289, 14490–14497.
- Pavankumar, T.L. Inteins: Localized distribution, gene regulation, and protein engineering for biological applications. Microorganisms 2018, 6, 19.
- Liu, X.-Q. Protein-splicing intein: Genetic mobility, origin, and evolution. Annu. Rev. Genet. 2000, 34, 61–76.
- di Ventura, B.; Mootz, H.D. Switchable inteins for conditional protein splicing. Biol. Chem. 2019, 400, 467–475.
- Buskirk, A.R.; Ong, Y.-C.; Gartner, Z.J.; Liu, D.R. Directed evolution of ligand dependence: Small-molecule-activated protein splicing. Proc. Natl. Acad. Sci. USA 2004, 101, 10505–10510.
- Peck, S.H.; Chen, I.; Liu, D.R. Directed evolution of a small-molecule-triggered intein with improved splicing properties in mammalian cells. Chem. Biol. 2011, 18, 619–630.
- Tan, G.; Chen, M.; Foote, C.; Tan, C. Temperature-sensitive mutations made easy: Generating conditional mutations by using temperature-sensitive inteins that function within different temperature ranges. Genetics 2009, 183, 13–22.
- Topilina, N.I.; Novikova, O.; Stanger, M.; Banavali, N.K.; Belfort, M. Post-translational environmental switch of RadA activity by extein–intein interactions in protein splicing. Nucleic Acids Res. 2015, 43, 6631–6648.
- Wood, D.W.; Wu, W.; Belfort, G.; Derbyshire, V.; Belfort, M. A genetic system yields self-cleaving inteins for bioseparations. Nat. Biotechnol. 1999, 17, 889–892.
- Chan, H.; Pearson, C.S.; Green, C.M.; Li, Z.; Zhang, J.; Belfort, G.; Shekhtman, A.; Li, H.; Belfort, M. Exploring intein inhibition by platinum compounds as an antimicrobial strategy. J. Biol. Chem. 2016, 291, 22661–22670.
- Liu, X.-Q.; Yang, J. Prp8 intein in fungal pathogens: Target for potential antifungal drugs. FEBS Lett. 2004, 572, 46–50.
- Paulus, H. Inteins as targets for potential antimycobacterial drugs. Front. Biosci. 2003, 8, S1157–S1165.
- Stevens, A.J.; Sekar, G.; Shah, N.H.; Mostafavi, A.Z.; Cowburn, D.; Muir, T.W. A promiscuous split intein with expanded protein engineering applications. Proc. Natl. Acad. Sci. USA 2017, 114, 8538–8543.
- Wood, D.W.; Camarero, J.A. Intein applications: From protein purification and labeling to metabolic control methods. J. Biol. Chem. 2014, 289, 14512–14519.
- Zhang, L.; Zheng, Y.; Callahan, B.; Belfort, M.; Liu, Y. Cisplatin inhibits protein splicing, suggesting inteins as therapeutic targets in mycobacteria. J. Biol. Chem. 2011, 286, 1277–1282.
- Hirata, R.; Ohsumk, Y.; Nakano, A.; Kawasaki, H.; Suzuki, K.; Anraku, Y. Molecular structure of a gene, VMA1, encoding the catalytic subunit of H (+)-translocating adenosine triphosphatase from vacuolar membranes of Saccharomyces cerevisiae. J. Biol. Chem. 1990, 265, 6726–6733.
- Kane, P.M.; Yamashiro, C.T.; Wolczyk, D.F.; Neff, N.; Goebl, M.; Stevens, T.H. Protein splicing converts the yeast TFP1 gene product to the 69-kD subunit of the vacuolar H (+)-adenosine triphosphatase. Science 1990, 250, 651–657.
- Hodges, R.A.; Perler, F.B.; Noren, C.J.; Jack, W.E. Protein splicing removes intervening sequences in an archaea DNA polymerase. Nucleic Acids Res. 1992, 20, 6153–6157.
- Davis, E.O.; Jenner, P.J.; Brooks, P.C.; Colston, M.J.; Sedgwick, S.G. Protein splicing in the maturation of M. tuberculosis recA protein: A mechanism for tolerating a novel class of intervening sequence. Cell 1992, 71, 201–210.
- Davis, E.O.; Sedgwick, S.G.; Colston, M.J. Novel structure of the recA locus of Mycobacterium tuberculosis implies processing of the gene product. J. Bacteriol. 1991, 173, 5653–5662.
- Davis, E.O.; Thangaraj, H.S.; Brooks, P.C.; Colston, M.J. Evidence of selection for protein introns in the recAs of pathogenic mycobacteria. Embo J. 1994, 13, 699–703.
- Gu, H.; Xu, J.; Gallagher, M.; Dean, G. Peptide splicing in the vacuolar ATPase subunit A from Candida tropicalis. J. Biol. Chem. 1993, 268, 7372–7381.
- Green, C.M.; Novikova, O.; Belfort, M. The dynamic intein landscape of eukaryotes. Mob. DNA 2018, 9, 4.
- Kelley, D.S.; Lennon, C.W.; Belfort, M.; Novikova, O. Mycobacteriophages as incubators for intein dissemination and evolution. MBio 2016, 7, e01537-16.
- Butler, M.I.; Gray, J.; Goodwin, T.J.; Poulter, R.T. The distribution and evolutionary history of the PRP8 intein. Bmc Evol. Biol. 2006, 6, 42.
- Elleuche, S.; Pöggeler, S. Fungal inteins: Distribution, evolution, and applications. In Physiology and Genetics; Springer: Berlin/Heidelberg, Germany, 2018; pp. 57–85.
- Butler, M.I.; Goodwin, T.J.; Poulter, R.T.M. A nuclear-encoded intein in the fungal pathogen Cryptococcus neoformans. Yeast 2001, 18, 1365–1370.
- Frederick, R.D.; Snyder, K.E.; Tooley, P.W.; Berthier-Schaad, Y.; Peterson, G.L.; Bonde, M.R.; Schaad, N.W.; Knorr, D.A. Identification and differentiation of Tilletia indica and T. walkeri using the polymerase chain reaction. Phytopathology 2000, 90, 951–960.
- Aherfi, S.; Colson, P.; La Scola, B.; Raoult, D. Giant viruses of amoebas: An update. Front. Microbiol. 2016, 7, 349.
- Dalgaard, J.Z.; Moser, M.J.; Hughey, R.; Mian, I.S. Statistical modeling, phylogenetic analysis and structure prediction of a protein splicing domain common to inteins and hedgehog proteins. J. Comput. Biol. 1997, 4, 193–214.
- Kawasaki, M.; Nogami, S.; Satow, Y.; Ohya, Y.; Anraku, Y. Identification of three core regions essential for protein splicing of the yeast VMA1 protozyme a random mutagenesis study of the entire Vma1-derived endonuclease sequence. J. Biol. Chem. 1997, 272, 15668–15674.
- Wu, H.; Hu, Z.; Liu, X.-Q. Protein trans-splicing by a split intein encoded in a split DnaE gene of Synechocystis sp. PCC6803. Proc. Natl. Acad. Sci. USA 1998, 95, 9226–9231.