
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Amancio Carnero | + 2308 word(s) | 2308 | 2021-08-18 11:38:08 | | | |
| 2 | Beatrix Zheng | + 395 word(s) | 2703 | 2021-08-19 05:33:55 | | |
Video Upload Options
Ovarian cancer (OC) is a gynecological malignancy responsible for 4.4% of cancer-related deaths in the world and is the most lethal gynecological tumor. The five-year overall survival of epithelial OC patients ranges from 20% at stage IV to 89% at stage I, but 80% of patients are diagnosed at advanced stages (III or IV) due to unspecific clinical manifestations. The standard treatment for OC consists of cytoreductive surgery combined with chemotherapy based on platinum (cisplatin or carboplatin) alone or in combination with paclitaxel, doxorubicin, or docetaxel, as well as poly (ADP-ribose) polymerase (PARP) inhibitors for patients with mutations in BRCA1 or BRCA2.
1. Introduction
Ovarian cancer (OC) is a gynecological malignancy responsible for 4.4% of cancer-related deaths in the world and is the most lethal gynecological tumor [1]. The five-year overall survival of epithelial OC patients ranges from 20% at stage IV to 89% at stage I, but 80% of patients are diagnosed at advanced stages (III or IV) due to unspecific clinical manifestations [2]. The standard treatment for OC consists of cytoreductive surgery combined with chemotherapy based on platinum (cisplatin or carboplatin) alone or in combination with paclitaxel, doxorubicin, or docetaxel, as well as poly (ADP-ribose) polymerase (PARP) inhibitors for patients with mutations in BRCA1 or BRCA2 [3]. Adjuvant intraperitoneal chemotherapy is also part of the standard treatment due to the poor drug distribution to the intraperitoneal cavity [4]. Histologically, there are three major OC types: epithelial tumors, which comprises 90% of cases; germ cell tumors, comprising 3% of cases; and sex cord-stromal tumors, comprising 2% of cases; with the remaining 5% of tumors’ type not specified ( Table 1 ) [2]. Epithelial OCs comprise four subtypes: serous, which could be high-grade serous (HG-SOC) or low-grade serous (LG-SOC); endometroid; clear cell; and mucinous [5]. While type I tumors, comprising LG-SOC, endometroid, clear cell, and mucinous carcinoma, as well as nonepithelial tumors, show less malignancy and genome instability, type II tumors comprise HG-SOC and are highly malignant and genetically unstable ( Table 1 ) [6]. HG-SOC constitutes 75% of total OC cases and therefore has been extensively studied.
| Histology | Epithelial Subtypes | Type | Clinical Features | Molecular Features | Stages at Detection | 5-Year OS |
|---|---|---|---|---|---|---|
| Epithelial (90%) |
High-grade serous (70–80%) | Type II | High grade, aggressive, bilateral, disseminated to omentum and peritoneum | GIN, ubiquitous mutations in TP53 and frequent in BRCA1/2, HR deficiency | Stage III–IV | 43% |
| Low-grade serous (<5%) |
Type I | Low grade (except clear cell), indolent, unilateral, restricted to ovary at diagnosis | No GIN, mutations in KRAS, BRAF, ERBB2, PI3K, PTEN, ARID1A, β-catenin | Stage I–II | ||
| Endometroid (10–15%) |
Stage I–II | 82% | ||||
| Mucinous (3–4%) | Stage I–II | 71% | ||||
| Clear cell (10%) | Stage I–II | 66% | ||||
| Sex cord stromal cell (2%) | Indolent | Stage I–II | 88% | |||
| Germ cell tumors (3%) | Indolent | Stage I–II | 94% | |||
| Others or unspecified (5%) | ||||||
In the last twenty years, genome-wide technologies have considerably advanced our knowledge about the molecular features of human cancers, including OC. In particular, the development of next-generation sequencing (NGS) has revolutionized how we investigate many biological questions. The ability to inspect whole genomes, transcriptomes, or different aspects of the epigenome in a single experiment has allowed an unprecedented increase in our understanding of how tumorigenesis works and the mechanisms by which cancer cells proliferate and avoid cellular controls and anticancer treatments. In this review, we cover recent advances in the molecular biology of OC using genomics, transcriptomics, and epigenomics ( Figure 1 ), with a focus on its biological features and therapy resistance.
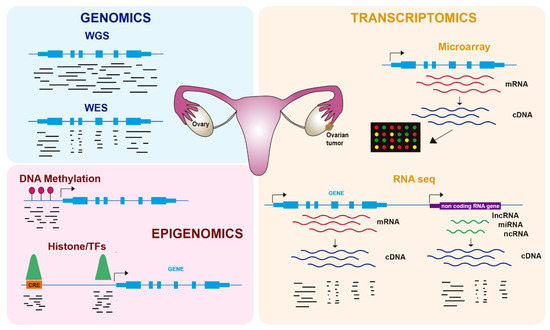
2. Genomics of OC
Several studies have used WES or WGS to identify genomic alterations involved in OC tumorigenesis, including somatic and germline mutations, copy number alterations (CNAs), and structural variants (SVs). The Cancer Genome Atlas (TCGA) study involving HG-SOC analyzed 489 patients’ mRNA and miRNA expression, promoter DNA methylation, and copy number, as well as WES in 316 of them and matched them with normal DNA samples [7]. They found TP53 mutations in virtually all tumors (96%), consistent with previous data [8], but other mutations were infrequent in HG-SOC. BRCA1 and BRCA2 showed germline mutations in 8 and 9% of cases, respectively, and somatic mutations in 3% of cases. Six additional genes showed statistically significant recurrent mutations in 2–6% of cases: RB1, NF1, FAT3, CSMD3, GABRA6, and CDK12. Finally, they described other rare mutations involving important HG-SOC drivers, such as BRAF, PIK3CA, KRAS, and NRAS. Furthermore, 113 CNAs were identified, including focal amplifications of CCNE1, MYC, MECOM, ZMYND8, IRF2BP2, ID4, PAX8, and TERT, as well as focal deletions of tumor suppressor genes such as PTEN, RB1, and NF1, the latter of which were also significantly mutated [7]. Integrative pathway analyses combining expression, methylation, and mutation showed altered pathways in HG-SOC, including RB1, PIK3/RAS, NOTCH, and FOXM1, and homologous recombination (HR) defects were also found for half of the analyzed tumors.
Subsequent studies in HG-SOC confirmed the main results of the TCGA study and identified new alterations. In this regard, Patch et al. used WGS together with transcriptome and methylome analyses in 92 patients [9]. They confirmed the prevalence of TP53 mutations in HG-SOC, as well as mutations in HR genes or BRCA1 promoter methylation in half of the tumors. They also identified gene breakage in tumor suppressor genes such as RB1, NF1, RAD51B, and PTEN, as well as CCNE1 amplification as the primary SV, consistent with previous results [10]. In addition to primary disease, this study also analyzed metastases to ascites for tumors sensitive to initial treatment but failing subsequent therapy. No clear relationship with chemoresistance was evident for the genes specifically mutated in the ascites tumoral cells; however, particular patients showed either reversions of BRCA1-2 mutations, similar to previous studies [11][12], loss of BRCA1 promoter methylation, or promoter fusions overexpressing the ABCB1 gene encoding the drug efflux transporter MDR1, which mediates rapid efflux of chemotherapeutic agents, including paclitaxel [13]. A larger study used WES with 2051 OC patients and found four susceptibility genes for OC: MSH6, RAD51C, TP53, and ATM [14], confirming the alterations in DNA repair found in OC.
Shallow whole-genome DNA sequencing was applied to 117 samples to study copy number signatures of HG-SOC [15]. This report described six copy number signatures underlying mutational processes in ovarian tumors occurring in parallel after widespread TP53 mutation. These signatures were associated with particular driver mutations and disease outcomes, including overall survival and tumor relapse after platinum treatment. For instance, copy number signature 3 is characterized by BRCA1-2-related HR defects and improved overall survival, consistent with previous data [16][17], while copy number signature 1 is characterized by oncogenic RAS signaling (mutations in NF1, KRAS, and NRAS) and predicts platinum-resistant relapse and poor survival.
Genomic techniques have also been used for studies on tumor evolution and heterogeneity. Several reports have analyzed the genomic landscape of OC metastases and reached the common conclusion that there is little accumulation of somatic mutations and CNAs in metastatic versus primary tumor samples. Lee et al. used 11 samples from primary and metastatic sites in a single HG-SOC patient [18]. By performing WES and sample clustering, they obtained three clusters: one of metastatic samples and two of primary tumor samples, where the metastatic samples arose from one of the primary tumor clusters with few additional somatic mutations and CNAs. Moreover, Schwarz et al. quantified the degree of tumor heterogeneity in 14 HG-SOC patients using WGS [19]. They found that a high degree of clonal expansion was associated with worse survival and that clonal populations detected at relapse originated in early branching events followed by divergent clonal evolution, which is consistent with previous observations showing that chemotherapy did not lead to substantial changes in genetic subclones and that genetically divergent subclones resistant to platinum may exist in the tumor before treatment [20].
3. Transcriptomics of OC
Gene expression was initially characterized in OC and other tumors using microarray-based technologies ( Figure 1 ), leading to the identification of several molecular subtypes. Tothill et al. profiled 285 serous and endometroid ovarian tumors and identified six clusters of ovarian tumors with distinct molecular features and malignant potential: two clusters of low-grade and four clusters of high-grade tumors [21]. These tumors included a novel high-grade serous subtype characterized by a mesenchymal cell state. In the TCGA study, 11,864 genes were analyzed by three different microarray-based platforms, leading to 1500 variable genes that were used to obtain four clusters: immunoreactive, differentiated, proliferative, and mesenchymal tumors [7]. Importantly, they identified a transcriptional signature formed by 193 genes, 108 of which were correlated with poor patient survival and 85 with good survival.
The origin of ovarian tumors in FTE [22] has been supported by studies based on RNA-seq. Qiu et al. analyzed gene expression in 31 tissue samples, including HG-SOC, LG-SOC, FTE, ovarian surface epithelium, and peritoneal mesothelia. They found that LG-SOC and HG-SOC samples clustered together with FTE and showed consistent overexpression of markers such as PAX8, CDH1, and FOXA2, suggesting a clonal origin of both HG-SOC and LG-SOC in FTE [23]. Indeed, a study using 102 human cancer cell lines previously identified PAX8 as differentially expressed in OC cell lines [24]. PAX8 is amplified in 16% of HG-SOC patients and overexpressed in ovarian tumors, suggesting that PAX8 is a lineage-specific essential gene, at least in a subset of OC cases. More recently, the transcription factor SOX18 has also been involved in the development of ovarian tumors from FTE [25].
The only OC biomarker currently approved for clinical use in early detection is circulating cancer antigen 125 (CA125), although it is common to other tumors, and it is only overexpressed in a subset of early stage tumors [26]. Therefore, there is a necessity to identify more specific and early expressed biomarkers of OC detection. In this regard, insulin-like growth factor binding protein 4 (IGFBP-4) was identified as upregulated in serous OC samples using RNA-seq, including early stage patients in which CA125 levels were normal [27]. Moreover, IGFBP-4 levels were higher in malignant than in benign disease. Another study analyzed the involvement of the solute carrier (SLC) family in OC patients, identifying SLC7A2 as downregulated and associated with poor prognosis [28]. Finally, Fei et al. analyzed published OC patient data and found that KIF11, CDC20, and TOPA were upregulated in ovarian tumors and correlated with worse patient survival [29].
Other studies have characterized the microenvironment of ovarian tumors, focusing on their degree of infiltration by immune cells. Olalekan et al. analyzed metastatic OC samples from the omentum of six patients and found cancer, stromal and immune cell types [30]. Two groups of samples were identified according to their high or low level of T cell infiltration, and their molecular characterization suggested an antitumor response in the highly infiltrated tumors. A very recent report characterized 15 ovarian tumors with different levels of immune infiltration that differed in their cell type composition [31]. They found chemokine receptor-ligand interactions as a potential mechanism driving immune cell infiltration. Both reports have major implications for immunotherapy in OC. Finally, Nath et al. investigated HG-SOC tumor evolution during chemotherapy using scRNA-seq and WGS of malignant ascites and pleural effusions [32]. They defined several transcriptional signatures or archetypes and found one of them characterized by high metabolic activity and proliferation, while there were no consistent genomic alterations defining those states. Interestingly, this metabolic and proliferative archetype was progressively enriched as chemotherapy progressed, and resistance to different lines of treatments was acquired.
4. Epigenomics of OC
Gene expression is regulated by processes that do not alter the DNA sequence and that are included under the term epigenetics. Epigenomics, which refers to the epigenetics of the whole genome, comprises reversible modifications such as DNA methylation or histone modifications (e.g., acetylation, methylation) ( Figure 1 ), as well as regulation by ncRNAs such as miRNAs. Tumors frequently use epigenetic alterations to proliferate and escape anticancer treatments, and therefore therapies targeting the epigenome are being increasingly tested [33]. In OC, epigenetic alterations, including DNA methylation and histone modifications, have been described in recent years as important contributors to tumorigenesis and chemoresistance [34] ( Table 2 ).
| Marker | Approach | Alteration | Treatment | Outcome | OC Subtype | Refs. |
|---|---|---|---|---|---|---|
| BRCA1, BRCA2 | WGS | Mutation reversion | Platinum | Resistance | HG-SOC | [9] |
| ADAMTS Family |
WES | Mutation | Platinum | Sensitivity | HG-SOC | [35] |
| TMEM205, POLR2A |
WES | Mutation | Paclitaxel + Carboplatin | Resistance | HG-SOC | [36] |
| PDL-L1 | WES | Structural variant | Pembrolizumab | Sensitivity | HG-SOC | [37] |
| KRAS | WES, WGS | Mutation | MEK inhibitors | Sensitivity | LG-SOC | [38] |
| MDR1 | RNA-seq | Up-regulated expression | Platinum | Resistance | HG-SOC | [9] |
| SAP25 HLA-DPA1 AKT3 PIK3R5 |
RNA-seq | Differential expression | Paclitaxel + Carboplatin | Resistance | HG-SOC | [36] |
| IRF1 | RNA-seq | Up-regulated expression | Platinum | Sensitivity | HG-SOC | [39] |
| DUOXA1 | RNA-seq | Up-regulated expression | Platinum | Resistance | Ovarian cell line | [40] |
| miR-137 | RNA-seq | Down-regulated expression | Cisplatin | Resistance | Ovarian cell line | [41] |
| HIF1A-AS2 | Bru-seq | Up-regulated expression | Olaparid + carboplatin + cisplatin | Sensitivity | Ovarian cell line | [42] |
| BRCA1 | Bisulfite chip | Loss of promoter methylation | Platinum | Resistance | HG-SOC | [9] |
| H3K27me3/H3K4me3 | ChIP-seq, RNA-seq |
Down-regulated expression | Platinum | Resistance | HG-SOC | [43] |
| H3K79me | ChIP-seq | Increased deposition | Platinum | Resistance | Ovarian cell line | [44] |
| SOX9 | ChIP-seq RNA-seq |
Up-regulated expression by superenhancers | Cisplatin | Resistance | Ovarian cell line | [45] |
| ISL1 | ChIP-seq RNA-seq |
Down-regulated expression by superenhancers | Cisplatin | resistance | Ovarian cell line | [46] |
DNA methylation at carbon 5 of cytosines (5-methylcytosine, 5mC) is an important player in the regulation of gene expression that typically occurs at CpG dinucleotides of gene promoters [47] ( Figure 1 ). Promoter DNA methylation is associated with gene repression and is regulated by DNA methyltransferases (DNMTs), which mediate de novo or maintenance methylation, and ten-eleven translocation (TET) proteins, which convert 5mC to 5-hydroxymethylcytosine (5hmC). In OC, as well as other types of tumors, two main methylation patterns have been found: hypermethylation of the promoters of tumor suppressor genes and hypomethylation of repeated sequences [48].
Finally, global loss of 5hmC levels has been associated with resistance to platinum-based therapy and worse patient survival [49]. Both overexpression of TET2 and treatment with the DNMT inhibitor 5-azacytidine, which in turn enhances TET gene expression, increase global 5hmC levels and restore platinum chemosensitivity, suggesting the use of DNMT inhibitors in ovarian tumors with low 5hmC levels. Another DNMT inhibitor, guadecitabine, was used in combination with carboplatin in OC patients, showing a higher sensitivity to chemotherapy than carboplatin alone and better patient survival [50]. Although drugs targeting DNA methylation represent a promising strategy to sensitize ovarian tumors to chemotherapy, the broad effect of these compounds represents a challenge that requires precise approaches to avoid undesirable effects. On the other hand, loss of methylation specifically at RAD51C promoter, whose methylation and subsequent silencing is related to HR defects, has been recently shown to cause resistance to PARP inhibitor treatment in HG-SOC PDX models [51].
Another layer of gene regulation is provided by cis-regulatory elements (CREs), which are DNA sequences harboring transcription factor (TF) binding sites that control the expression of target genes [52]. CREs are basically composed of promoters and enhancers that act at long distances over target promoters, likely by a looping mechanism. Histone modifications at these CREs define their activation status and accessibility, which is due to the binding of TFs. The most commonly used ‘omics’ approach to inspect either histone modifications or TF binding genome wide is chromatin immunoprecipitation coupled to sequencing (ChIP-seq), which employs specific antibodies to isolate and sequence the DNA associated with these proteins ( Figure 1 ).
5. Concluding Remarks
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424.
- Torre, L.A.; Trabert, B.; DeSantis, C.E.; Miller, K.D.; Samimi, G.; Runowicz, C.D.; Gaudet, M.M.; Jemal, A.; Siegel, R.L. Ovarian cancer statistics, 2018. CA Cancer J. Clin. 2018, 68, 284–296.
- Pignata, S.; Cannella, L.; Leopardo, D.; Pisano, C.; Bruni, G.S.; Facchini, G. Chemotherapy in epithelial ovarian cancer. Cancer Lett. 2011, 303, 73–83.
- Armstrong, D.K.; Bundy, B.; Wenzel, L.; Huang, H.Q.; Baergen, R.; Lele, S.; Copeland, L.J.; Walker, J.L.; Burger, R.A. Intraperitoneal cisplatin and paclitaxel in ovarian cancer. N. Engl. J. Med. 2006, 354, 34–43.
- Mallen, A.; Soong, T.R.; Townsend, M.K.; Wenham, R.M.; Crum, C.P.; Tworoger, S.S. Surgical prevention strategies in ovarian cancer. Gynecol. Oncol. 2018, 151, 166–175.
- Kurman, R.J.; Shih, I.M. The Dualistic Model of Ovarian Carcinogenesis: Revisited, Revised, and Expanded. Am. J. Pathol. 2016, 186, 733–747.
- Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 2011, 474, 609–615.
- Ahmed, A.A.; Etemadmoghadam, D.; Temple, J.; Lynch, A.G.; Riad, M.; Sharma, R.; Stewart, C.; Fereday, S.; Caldas, C.; Defazio, A.; et al. Driver mutations in TP53 are ubiquitous in high grade serous carcinoma of the ovary. J. Pathol. 2010, 221, 49–56.
- Patch, A.M.; Christie, E.L.; Etemadmoghadam, D.; Garsed, D.W.; George, J.; Fereday, S.; Nones, K.; Cowin, P.; Alsop, K.; Bailey, P.J.; et al. Whole-genome characterization of chemoresistant ovarian cancer. Nature 2015, 521, 489–494.
- Etemadmoghadam, D.; deFazio, A.; Beroukhim, R.; Mermel, C.; George, J.; Getz, G.; Tothill, R.; Okamoto, A.; Raeder, M.B.; Harnett, P.; et al. Integrated genome-wide DNA copy number and expression analysis identifies distinct mechanisms of primary chemoresistance in ovarian carcinomas. Clin. Cancer Res. 2009, 15, 1417–1427.
- Norquist, B.; Wurz, K.A.; Pennil, C.C.; Garcia, R.; Gross, J.; Sakai, W.; Karlan, B.Y.; Taniguchi, T.; Swisher, E.M. Secondary somatic mutations restoring BRCA1/2 predict chemotherapy resistance in hereditary ovarian carcinomas. J. Clin. Oncol. 2011, 29, 3008–3015.
- Sakai, W.; Swisher, E.M.; Karlan, B.Y.; Agarwal, M.K.; Higgins, J.; Friedman, C.; Villegas, E.; Jacquemont, C.; Farrugia, D.J.; Couch, F.J.; et al. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature 2008, 451, 1116–1120.
- Murray, S.; Briasoulis, E.; Linardou, H.; Bafaloukos, D.; Papadimitriou, C. Taxane resistance in breast cancer: Mechanisms, predictive biomarkers and circumvention strategies. Cancer Treat. Rev. 2012, 38, 890–903.
- Lu, H.-M.; Li, S.; Black, M.H.; Lee, S.; Hoiness, R.; Wu, S.; Mu, W.; Huether, R.; Chen, J.; Sridhar, S.; et al. Association of Breast and Ovarian Cancers with Predisposition Genes Identified by Large-Scale Sequencing. JAMA Oncol. 2019, 5, 51–57.
- Macintyre, G.; Goranova, T.E.; De Silva, D.; Ennis, D.; Piskorz, A.M.; Eldridge, M.; Sie, D.; Lewsley, L.-A.; Hanif, A.; Wilson, C.; et al. Copy number signatures and mutational processes in ovarian carcinoma. Nat. Genet. 2018, 50, 1262–1270.
- dos Reis, F.J.C.; Song, H.; Goode, E.L.; Cunningham, J.M.; Fridley, B.L.; Larson, M.C.; Alsop, K.; Dicks, E.; Harrington, P.; Ramus, S.J.; et al. Germline mutation in BRCA1 or BRCA2 and ten-year survival for women diagnosed with epithelial ovarian cancer. Clin. Cancer Res. 2015, 21, 652–657.
- Norquist, B.M.; Brady, M.F.; Harrell, M.I.; Walsh, T.; Lee, M.K.; Gulsuner, S.; Bernards, S.S.; Casadei, S.; Burger, R.A.; Tewari, K.S.; et al. Mutations in Homologous Recombination Genes and Outcomes in Ovarian Carcinoma Patients in GOG 218: An NRG Oncology/Gynecologic Oncology Group Study. Clin. Cancer Res. 2018, 24, 777–783.
- Lee, J.-Y.; Yoon, J.-K.; Kim, B.; Kim, S.; Kim, M.-A.; Lim, H.; Bang, D.; Song, Y.-S. Tumor evolution and intratumor heterogeneity of an epithelial ovarian cancer investigated using next-generation sequencing. BMC Cancer 2015, 15, 85.
- Schwarz, R.F.; Ng, C.K.; Cooke, S.L.; Newman, S.; Temple, J.; Piskorz, A.M.; Gale, D.; Sayal, K.; Murtaza, M.; Baldwin, P.J.; et al. Spatial and temporal heterogeneity in high-grade serous ovarian cancer: A phylogenetic analysis. PLoS Med. 2015, 12, e1001789.
- Cooke, S.L.; Ng, C.K.; Melnyk, N.; Garcia, M.J.; Hardcastle, T.; Temple, J.; Langdon, S.; Huntsman, D.; Brenton, J.D. Genomic analysis of genetic heterogeneity and evolution in high-grade serous ovarian carcinoma. Oncogene 2010, 29, 4905–4913.
- Tothill, R.W.; Tinker, A.V.; George, J.; Brown, R.; Fox, S.B.; Lade, S.; Johnson, D.S.; Trivett, M.K.; Etemadmoghadam, D.; Locandro, B.; et al. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin. Cancer Res. 2008, 14, 5198–5208.
- Karnezis, A.N.; Cho, K.R.; Gilks, C.B.; Pearce, C.L.; Huntsman, D.G. The disparate origins of ovarian cancers: Pathogenesis and prevention strategies. Nat. Rev. Cancer 2017, 17, 65–74.
- Qiu, C.; Lu, N.; Wang, X.; Zhang, Q.; Yuan, C.; Yan, S.; Dongol, S.; Li, Y.; Sun, X.; Sun, C.; et al. Gene expression profiles of ovarian low-grade serous carcinoma resemble those of fallopian tube epithelium. Gynecol. Oncol. 2017, 147, 634–641.
- Cheung, H.W.; Cowley, G.S.; Weir, B.A.; Boehm, J.S.; Rusin, S.; Scott, J.A.; East, A.; Ali, L.D.; Lizotte, P.H.; Wong, T.C.; et al. Systematic investigation of genetic vulnerabilities across cancer cell lines reveals lineage-specific dependencies in ovarian cancer. Proc. Natl. Acad. Sci. USA 2011, 108, 12372–12377.
- Lawrenson, K.; Fonseca, M.A.S.; Liu, A.Y.; Dezem, F.S.; Lee, J.M.; Lin, X.; Corona, R.I.; Abbasi, F.; Vavra, K.C.; Dinh, H.Q.; et al. A Study of High-Grade Serous Ovarian Cancer Origins Implicates the SOX18 Transcription Factor in Tumor Development. Cell Rep. 2019, 29, 3726–3735.e4.
- Felder, M.; Kapur, A.; Gonzalez-Bosquet, J.; Horibata, S.; Heintz, J.; Albrecht, R.; Fass, L.; Kaur, J.; Hu, K.; Shojaei, H.; et al. MUC16 (CA125): Tumor biomarker to cancer therapy, a work in progress. Mol. Cancer 2014, 13, 129.
- Mosig, R.A.; Lobl, M.; Senturk, E.; Shah, H.; Cohen, S.; Chudin, E.; Fruscio, R.; Marchini, S.; D’Incalci, M.; Sachidanandam, R.; et al. IGFBP-4 tumor and serum levels are increased across all stages of epithelial ovarian cancer. J. Ovarian Res. 2012, 5, 3.
- Sun, T.; Bi, F.; Liu, Z.; Yang, Q. SLC7A2 serves as a potential biomarker and therapeutic target for ovarian cancer. Aging 2020, 12, 13281–13296.
- Fei, H.; Chen, S.; Xu, C. Bioinformatics analysis of gene expression profile of serous ovarian carcinomas to screen key genes and pathways. J. Ovarian Res. 2020, 13, 82.
- Olalekan, S.; Xie, B.; Back, R.; Eckart, H.; Basu, A. Characterizing the tumor microenvironment of metastatic ovarian cancer by single-cell transcriptomics. Cell Rep. 2021, 35, 109165.
- Hornburg, M.; Desbois, M.; Lu, S.; Guan, Y.; Lo, A.A.; Kaufman, S.; Elrod, A.; Lotstein, A.; DesRochers, T.M.; Munoz-Rodriguez, J.L.; et al. Single-cell dissection of cellular components and interactions shaping the tumor immune phenotypes in ovarian cancer. Cancer Cell 2021, 39, 928–944.e6.
- Nath, A.; Cosgrove, P.A.; Mirsafian, H.; Christie, E.L.; Pflieger, L.; Copeland, B.; Majumdar, S.; Cristea, M.C.; Han, E.S.; Lee, S.J.; et al. Evolution of core archetypal phenotypes in progressive high grade serous ovarian cancer. Nat. Commun. 2021, 12, 3039.
- Jones, P.A.; Issa, J.-P.; Baylin, S. Targeting the cancer epigenome for therapy. Nat. Rev. Genet. 2016, 17, 630–641.
- Matei, D.; Nephew, K.P. Epigenetic Attire in Ovarian Cancer: The Emperor’s New Clothes. Cancer Res. 2020, 80, 3775–3785.
- Liu, Y.; Yasukawa, M.; Chen, K.; Hu, L.; Broaddus, R.R.; Ding, L.; Mardis, E.R.; Spellman, P.; Levine, D.A.; Mills, G.B.; et al. Association of Somatic Mutations of ADAMTS Genes with Chemotherapy Sensitivity and Survival in High-Grade Serous Ovarian Carcinoma. JAMA Oncol. 2015, 1, 486–494.
- Li, L.Y.; Kim, H.J.; Park, S.A.; Lee, S.H.; Kim, L.K.; Lee, J.Y.; Kim, S.; Kim, Y.T.; Kim, S.W.; Nam, E.J. Genetic Profiles Associated with Chemoresistance in Patient-Derived Xenograft Models of Ovarian Cancer. Cancer Res. Treat. 2019, 51, 1117–1127.
- Bellone, S.; Buza, N.; Choi, J.; Zammataro, L.; Gay, L.; Elvin, J.; Rimm, D.L.; Liu, Y.; Ratner, E.S.; Schwartz, P.E.; et al. Exceptional Response to Pembrolizumab in a Metastatic, Chemotherapy/Radiation-Resistant Ovarian Cancer Patient Harboring a PD-L1-Genetic Rearrangement. Clin. Cancer Res. 2018, 24, 3282–3291.
- Fernandez, M.L.; Dawson, A.; Hoenisch, J.; Kim, H.; Bamford, S.; Salamanca, C.; DiMattia, G.; Shepherd, T.; Cremona, M.; Hennessy, B.; et al. Markers of MEK inhibitor resistance in low-grade serous ovarian cancer: EGFR is a potential therapeutic target. Cancer Cell Int. 2019, 19, 10.
- Cohen, S.; Mosig, R.; Moshier, E.; Pereira, E.; Rahaman, J.; Prasad-Hayes, M.; Halpert, R.; Billaud, J.N.; Dottino, P.; Martignetti, J.A. Interferon regulatory factor 1 is an independent predictor of platinum resistance and survival in high-grade serous ovarian carcinoma. Gynecol. Oncol. 2014, 134, 591–598.
- Meng, Y.; Chen, C.W.; Yung, M.M.H.; Sun, W.; Sun, J.; Li, Z.; Li, J.; Li, Z.; Zhou, W.; Liu, S.S.; et al. DUOXA1-mediated ROS production promotes cisplatin resistance by activating ATR-Chk1 pathway in ovarian cancer. Cancer Lett. 2018, 428, 104–116.
- Sun, J.; Cai, X.; Yung, M.M.; Zhou, W.; Li, J.; Zhang, Y.; Li, Z.; Liu, S.S.; Cheung, A.N.Y.; Ngan, H.Y.S.; et al. miR-137 mediates the functional link between c-Myc and EZH2 that regulates cisplatin resistance in ovarian cancer. Oncogene 2019, 38, 564–580.
- Lu, T.; Tang, J.; Shrestha, B.; Heath, B.R.; Hong, L.; Lei, Y.L.; Ljungman, M.; Neamati, N. Up-regulation of hypoxia-inducible factor antisense as a novel approach to treat ovarian cancer. Theranostics 2020, 10, 6959–6976.
- Chapman-Rothe, N.; Curry, E.; Zeller, C.; Liber, D.; Stronach, E.; Gabra, H.; Ghaem-Maghami, S.; Brown, R. Chromatin H3K27me3/H3K4me3 histone marks define gene sets in high-grade serous ovarian cancer that distinguish malignant, tumour-sustaining and chemo-resistant ovarian tumour cells. Oncogene 2013, 32, 4586–4592.
- Liu, D.; Zhang, X.-X.; Li, M.-C.; Cao, C.-H.; Wan, D.-Y.; Xi, B.-X.; Tan, J.-H.; Wang, J.; Yang, Z.-Y.; Feng, X.-X.; et al. C/EBPbeta enhances platinum resistance of ovarian cancer cells by reprogramming H3K79 methylation. Nat. Commun. 2018, 9, 1739.
- Shang, S.; Yang, J.; Jazaeri, A.A.; Duval, A.J.; Tufan, T.; Fischer, N.L.; Benamar, M.; Guessous, F.; Lee, I.; Campbell, R.M.; et al. Chemotherapy-Induced Distal Enhancers Drive Transcriptional Programs to Maintain the Chemoresistant State in Ovarian Cancer. Cancer Res. 2019, 79, 4599–4611.
- Ma, Q.; Yang, F.; Mackintosh, C.; Jayani, R.S.; Oh, S.; Jin, C.; Nair, S.J.; Merkurjev, D.; Ma, W.; Allen, S.; et al. Super-Enhancer Redistribution as a Mechanism of Broad Gene Dysregulation in Repeatedly Drug-Treated Cancer Cells. Cell Rep. 2020, 31, 107532.
- Skvortsova, K.; Iovino, N.; Bogdanovic, O. Functions and mechanisms of epigenetic inheritance in animals. Nat. Rev. Mol. Cell Biol. 2018, 19, 774–790.
- Earp, M.A.; Cunningham, J.M. DNA methylation changes in epithelial ovarian cancer histotypes. Genomics 2015, 106, 311–321.
- Tucker, D.W.; Getchell, C.R.; McCarthy, E.T.; Ohman, A.W.; Sasamoto, N.; Xu, S.; Ko, J.Y.; Gupta, M.; Shafrir, A.; Medina, J.E.; et al. Epigenetic Reprogramming Strategies to Reverse Global Loss of 5-Hydroxymethylcytosine, a Prognostic Factor for Poor Survival in High-grade Serous Ovarian Cancer. Clin. Cancer Res. 2018, 24, 1389–1401.
- Fang, F.; Cardenas, H.; Huang, H.; Jiang, G.; Perkins, S.M.; Zhang, C.; Keer, H.N.; Liu, Y.; Nephew, K.P.; Matei, D. Genomic and Epigenomic Signatures in Ovarian Cancer Associated with Resensitization to Platinum Drugs. Cancer Res. 2018, 78, 631–644.
- Nesic, K.; Kondrashova, O.; Hurley, R.M.; McGehee, C.D.; Vandenberg, C.J.; Ho, G.Y.; Lieschke, E.; Dall, G.; Bound, N.; Shield-Artin, K.; et al. Acquired RAD51C promoter methylation loss causes PARP inhibitor resistance in high grade serous ovarian carcinoma. Cancer Res. 2021.
- Andersson, R.; Sandelin, A. Determinants of enhancer and promoter activities of regulatory elements. Nat. Rev. Genet. 2020, 21, 71–87.




