1000/1000
Hot
Most Recent

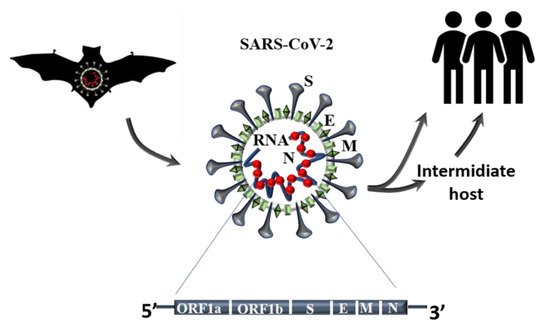
Since the beginning of 2020, the new pandemic caused by SARS-CoV-2 and named coronavirus disease 19 (COVID 19) has changed our socio-economic life. In just a few months, SARS-CoV-2 was able to spread worldwide at an unprecedented speed, causing hundreds of thousands of deaths, especially among the weakest part of the population. Indeed, especially at the beginning of this pandemic, many reports highlighted how people, suffering from other pathologies, such as hypertension, cardiovascular diseases, and diabetes, are more at risk of severe outcomes if infected. Although this pandemic has put the entire academic world to the test, it has also been a year of intense research and many important contributions have advanced our understanding of SARS-CoV-2 origin, its molecular structure and its mechanism of infection. Unfortunately, despite this great effort, we are still a long way from fully understanding how SARS-CoV-2 dysregulates organismal physiology and whether the current vaccines will be able to protect us from possible future pandemics.
| SARS-CoV-2 | Bat CoV | BtRs-BetaCoV/YN2018A | SARS -CoV | MERS | hCoV 229E |
hCoV HKU1 | hCoV NL63 | hCoV OC43 |
|---|---|---|---|---|---|---|---|---|
| 99% | 96% | 96% | 70% | 58% | 67% | 59% | 66% |
| Clinical Report | Nr Cases | Age | Males | Females | CVD | Diabetes | Hypertension |
|---|---|---|---|---|---|---|---|
| Zhonghua Liu Xing 2020 [38] | 44672 | 30–69 (77.8%) | 51.4% | 48.6% | 4.2% | 1.1% | 12.8% |
| Xie J et al., 2020 [39] | 168 | >50 | 75% | 25% | 18.5% | 25% | 50% |
| Guan WJ et al., 2020 [21] | 1099 | >50 (56%) | 58.1% | 41.9% | 2.5% | 7.4% | 15% |
| Huang C et al., 2020 [40] | 41 | 49 median | 73% | 27% | 15% | 20% | 15% |
| Zhang JJ et al., 2020 [41] | 140 | 57 median | 50.7% | 49.3% | not specified | 12.1% | 30% |
| Li Q et al., 2020 [42] | 425 | 59 median | 56% | 44% | not specified | ||
| Wang D et al., 2020 [43] | 138 | 56 median | 54.3% | 45.7% | 10.1% | 14.5% | 31.2% |
| Chen N et al., 2020 [44] | 99 | 55.5 median | 67.7% | 32.3% | 40% | 13% | 3% |
| Shi H et al., 2020 [45] | 81 | 49.5 median | 52% | 48% | 10% | 12% | 15% |
| Liang WH 2020 [46] | 1590 | 48.9 median | 57.3% | 42.7% | 3.7% | 8.2% | 16.9% |
| Group | Drugs Name | Action |
|---|---|---|
| Anti-Inflammatory | 1) Azithromycin | 1) Immuno-modulatory effect; |
| 2) Tocilizumab | 2) Humanized anti Il-6 receptor antibody, it bind soluble and membrane receptors blocking JAK-STAT pathways reducing inflammation; | |
| 3) Corticosteroids | 3) Helps dampens inflammation and other immune response; | |
| 4) Thalidomide | 4) Reduction of cytokine storm; | |
| 5) Anakinra | 5) Block IL-1; | |
| 6) Rituxolitinib | 6) JAK1 and JAK2 inhibitor; | |
| 7) Bacitinib | 7) Inhibits the kinase activities of JAK1 and JAK2. | |
| Anti-Viral | 1) Hydroxychloroquine; | 1) Inhibit the virus entry into host cells increasing endosomal pH resulting in inhibition of membrane fusion between host cell and virus; |
| 2) Camostat; | 2) Block viral maturation and entry into host cells; | |
| 3) Remdesivir; | 3) It terminates RNA synthesis and inhibits SARS-CoV-2 genome replication; | |
| 4) Lopinavir; | 4) Protease inhibitor, used in combination with ritonavir improving anti-viral activity; | |
| 5) Ritonavir; | 5) Used in combination with lopinavir; | |
| 6) Favipiravir; | 6) It is a guanine analogue, inhibits RNA polymerase; | |
| 7) Umifenovir; | 7) Inhibit viral and cellular membrane fusion; | |
| 8) Ivermectin. | 8) Block viral replication. | |
| Monoclonal Antibody | 1) Casirivimab; | 1) Block viral entry into host cell; |
| 2) Imdevimab; | 2) Block viral entry into host cell; | |
| 3) Bamlanivimab. | 3) Block viral entry into host cell. | |
| Plasma Therapy | Immune serum (convalescent plasma) | Exploitation of virus-specific antibody |
| Cell- Based Therapy | 1) Mesenchymal stem cell; | 1) Ameliorate tissue regeneration; |
| 2) Natural Killer cell | 2) Enhance immune response. |