
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Ivana Strazic Geljic | + 1135 word(s) | 1135 | 2021-06-03 05:34:19 | | | |
| 2 | Lily Guo | + 6 word(s) | 1141 | 2021-06-13 04:59:37 | | |
Video Upload Options
Numerous viruses hijack cellular protein trafficking pathways to mediate cell entry or to rearrange membrane structures thereby promoting viral replication and antagonizing the immune response. Adaptor protein complexes (AP), which mediate protein sorting in endocytic and secretory transport pathways, are one of the conserved viral targets. We present here different mechanisms of viral interference with AP complexes and the functional consequences that allow for efficient viral propagation and evasion of host immune defense. The best described examples are interactions of human immunodeficiency virus and human herpesviruses with AP complexes. Several other viruses, like Ebola, Nipah, and SARS-CoV-2, are pointed out as high priority disease-causative agents supporting the need for deeper understanding of virus-AP interplay which can be exploited in the design of novel antiviral therapies
1. Introduction
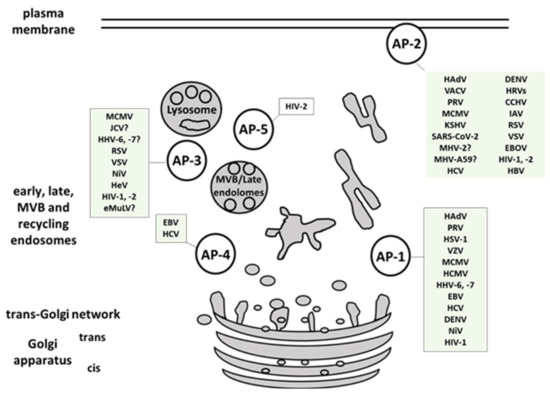
2. Viral Interactions with Adaptor-Protein Complexes
| Group | Viral Family Members included in the Review | Viral Protein Involved | AP Complex Involved | References | ||
|---|---|---|---|---|---|---|
| dsDNA Viruses | Adenoviridae | Human Adenoviruses (HAdV, HAdV2, HAdV5) | RIDα, RIDβ | AP-1, AP-2 | [11][12][13] | |
| Herpesviridae | α-herpesvirinae | Herpes simplex virus 1 and 2 (HSV-1, HSV-2) | gE/gI, VP22 | AP-1 | [14][15][16] | |
| Pseudorabiesvirus (PRV) | gB, gE | AP-1, AP-2 | [17] | |||
| Varicella zoster virus (VZV) | ORF9p | AP-1 | [18] | |||
| β-herpesvirinae | Murine Cytomegalovirus (MCMV) | m154, gp48, m04 | AP-1, AP-2, AP-3 | [19][20][21] | ||
| Human Cytomegalovirus (HCMV) | UL20 | AP-1 | [22] | |||
| Human herpes virus 6, -7 (HHV-6, -7) | U21 | AP-1, AP-3 | [23] | |||
| γ-herpesvirinae | Epstein-Barr virus (EBV) | BILF1, BMRF, | AP-1, AP-4 | [24][25] | ||
| Kaposi’s sarcoma-associated herpesvirus (KSHV) | unknown | AP-2 | [26] | |||
| Polyomaviridae | Human polyomavirus (JCV) | agnoprotein | AP-2, AP-3 | [27] | ||
| Poxviridae | Vaccinia virus (VACV) | VACV F13, VACV A33 | AP-2 | [28][29] | ||
| ssDNA viruses (+ strand or “sense”) | not identified | / | / | / | / | |
| dsRNA viruses | not identified | / | / | / | / | |
| (+) ssRNA viruses (+ strand or sense) |
Coronavirdae | Severe acute respiratory syndrome coronavirus (SARS-CoV, SARS-CoV-2) | unknown | AP-2 | [9][30][31] | |
| Murine hepatitis virus (MHV) | unknown | AP-2 | [32][33] | |||
| Flaviviridae | Dengue virus (DENV) | unknown | AP-1, AP-2 | [34][35] | ||
| Hepatitis C virus (HCV) | NS2, NS5A, core | AP-1, AP-2, AP-4 | [36][37][38][39][40][1] | |||
| Rhinoviridae | Human rhinovirus (hRV) | LDLR (minor group) | AP-2 | [41] | ||
| (−) ssRNA viruses (− strand or antisense) RNA | Filoviridae | Zaire ebolavirus (EBOV) | unknown | AP-1, AP-2 | [42][43] | |
| Nairoviriade | Crimean-congo hemorragic fever (CCHFV) | unknown | AP-2 | [44] | ||
| Orthomyxoviridae | Influenza A (IAV) | Hemagglutinin | AP-2 | [9][45][46] | ||
| Paramyxoviridae | Hendra virus (HeV) | M-protein | AP-3 | [47] | ||
| Nipah virus (NiV) | NiV-F, M-protein | AP-1, AP-3 | [47][48] | |||
| Pneumoviridae | Human respiratory syncytial virus (RSV) | M-protein | AP-2, AP-3 | [49][50][51] | ||
| Rhabdoviridae | Vesicular stomatitis Indiana virus (VSV) | VSV-G | AP-2, AP-3 | [52][53] | ||
| ssRNA-RT viruses (+ strand or sense) RNA with DNA intermediate in life-cycle | Retroviridae | Ecotropic murine leukemia virus (eMuLV) | Env | AP-3? | [54] | |
| Human immunodeficiency virus 1 (HIV-1) | Nef, Gag, Vpu | AP-1, AP-2, AP-3 | [55][56][57][58] | |||
| Human immunodeficiency virus 2 (HIV-2) | Nef, Env, Gag | AP-1, AP-2, AP-3, AP-5 | [59][60][61] | |||
| dsDNA-RT viruses DNA with RNA intermediate in life-cycle | Hepadnaviridae | Hepatitis B virus (HVB) | preS1 | AP-2 | [62][63][64] | |
References
- Robinson, M.; Schor, S.; Barouch-Bentov, R.; Einav, S. Viral journeys on the intracellular highways. Cell Mol Life Sci. 2018, 75, 3693–3714.
- Ren, S.; Ding, C.; Sun, Y. Morphology Remodeling and Selective Autophagy of Intracellular Organelles during Viral Infections. Int. J. Mol. Sci. 2020, 21, 3689.
- Agrawal, T.; Schu, P.; Medigeshi, G.R. Adaptor protein complexes-1 and 3 are involved at distinct stages of flavivirus life-cycle. Sci. Rep. 2013, 3, 1813.
- Boehm, M.; Bonifacino, J.S. Adaptins. Mol. Biol. Cell 2001, 12, 2907–2920.
- Hirst, J.; D. Barlow, L.; Francisco, G.C.; Sahlender, D.A.; Seaman, M.N.J.; Dacks, J.B.; Robinson, M.S. The Fifth Adaptor Protein Complex. PLoS Biol. 2011, 9, e1001170.
- Robinson, M.S.; Bonifacino, J.S. Adaptor-related proteins. Curr. Opin. Cell Biol. 2001, 13, 444–453.
- Hirst, J.; Itzhak, D.N.; Antrobus, R.; Borner, G.H.H.; Robinson, M.S. Role of the AP-5 adaptor protein complex in late endosome-to-Golgi retrieval. PLoS Biol. 2018, 16, e2004411.
- Brodsky, F.M. Diversity of clathrin function: New tricks for an old protein. Annu. Rev. Cell Dev. Biol. 2012, 28, 309–336.
- Yuan, S.; Chu, H.; Huang, J.; Zhao, X.; Ye, Z.W.; Lai, P.M.; Wen, L.; Cai, J.P.; Mo, Y.; Cao, J.; et al. Viruses harness YxxO motif to interact with host AP2M1 for replication: A vulnerable broad-spectrum antiviral target. Sci. Adv. 2020, 6, eaba7910.
- Helenius, A.; Kartenbeck, J.; Simons, K.; Fries, E. On the entry of Semliki forest virus into BHK-21 cells. J. Cell Biol. 1980, 84, 404–420.
- Carvajal-Gonzalez, J.M.; Gravotta, D.; Mattera, R.; Diaz, F.; Perez Bay, A.; Roman, A.C.; Schreiner, R.P.; Thuenauer, R.; Bonifacino, J.S.; Rodriguez-Boulan, E. Basolateral sorting of the coxsackie and adenovirus receptor through interaction of a canonical YXXPhi motif with the clathrin adaptors AP-1A and AP-1B. Proc. Natl. Acad. Sci. USA 2012, 109, 3820–3825.
- Cianciola, N.L.; Crooks, D.; Shah, A.H.; Carlin, C. A tyrosine-based signal plays a critical role in the targeting and function of adenovirus RIDalpha protein. J. Virol. 2007, 81, 10437–10450.
- Flatt, J.W.; Butcher, S.J. Adenovirus flow in host cell networks. Open Biol. 2019, 9, 190012.
- DNAM-1 and PVR regulate monocyte migration through endothelial junctions. J. Exp. Med. 2004, 199, 1331–1341.
- Johnson, D.C.; Webb, M.; Wisner, T.W.; Brunetti, C. Herpes Simplex Virus gE/gI Sorts Nascent Virions to Epithelial Cell Junctions, Promoting Virus Spread. J. Virol. 2001, 75, 821–833.
- Liu, J.; Gallo, R.M.; Duffy, C.; Brutkiewicz, R.R. A VP22-Null HSV-1 Is Impaired in Inhibiting CD1d-Mediated Antigen Presentation. Viral Immunol. 2016, 29, 409–416.
- Van Minnebruggen, G.; Favoreel, H.W.; Nauwynck, H.J. Internalization of Pseudorabies Virus Glycoprotein B Is Mediated by an Interaction between the YQRL Motif in Its Cytoplasmic Domain and the Clathrin-Associated AP-2 Adaptor Complex. J. Virol. 2004, 78, 8852–8859.
- Lebrun, M.; Lambert, J.; Riva, L.; Thelen, N.; Rambout, X.; Blondeau, C.; Thiry, M.; Snoeck, R.; Twizere, J.-C.; Dequiedt, F.; et al. Varicella-Zoster Virus ORF9p Binding to Cellular Adaptor Protein Complex 1 Is Important for Viral Infectivity. J. Virol. 2018, 92, e00295-18.
- Reusch, U.; Bernhard, O.; Koszinowski, U.; Schu, P. AP-1A and AP-3A Lysosomal Sorting Functions. Traffic 2002, 3, 752–761.
- Strazic Geljic, I.; Kucan Brlic, P.; Angulo, G.; Brizic, I.; Lisnic, B.; Jenus, T.; Juranic Lisnic, V.; Pietri, G.P.; Engel, P.; Kaynan, N.; et al. Cytomegalovirus protein m154 perturbs the adaptor protein-1 compartment mediating broad-spectrum immune evasion. eLife 2020, 9, e50803.
- Fink, A.; Blaum, F.; Babic Cac, M.; Ebert, S.; Lemmermann, N.A.; Reddehase, M.J. An endocytic YXXPhi (YRRF) cargo sorting motif in the cytoplasmic tail of murine cytomegalovirus AP2 ’adapter adapter’ protein m04/gp34 antagonizes virus evasion of natural killer cells. Med. Microbiol. Immunol. 2015, 204, 383–394.
- Jelcic, I.; Reichel, J.; Schlude, C.; Treutler, E.; Sinzger, C.; Steinle, A. The Polymorphic HCMV Glycoprotein UL20 Is Targeted for Lysosomal Degradation by Multiple Cytoplasmic Dileucine Motifs. Traffic 2011, 12, 1444–1456.
- Kimpler, L.A.; Glosson, N.L.; Downs, D.; Gonyo, P.; May, N.A.; Hudson, A.W. Adaptor Protein Complexes AP-1 and AP-3 Are Required by the HHV-7 Immunoevasin U21 for Rerouting of Class I MHC Molecules to the Lysosomal Compartment. PLoS ONE 2014, 9, e99139.
- Xiao, J.; Palefsky, J.M.; Herrera, R.; Berline, J.; Tugizov, S.M. EBV BMRF-2 facilitates cell-to-cell spread of virus within polarized oral epithelial cells. Virology 2009, 388, 335–343.
- Zuo, J.; Quinn, L.L.; Tamblyn, J.; Thomas, W.A.; Feederle, R.; Delecluse, H.-J.; Hislop, A.D.; Rowe, M. The Epstein-Barr Virus-Encoded BILF1 Protein Modulates Immune Recognition of Endogenously Processed Antigen by Targeting Major Histocompatibility Complex Class I Molecules Trafficking on both the Exocytic and Endocytic Pathways. J. Virol. 2011, 85, 1604–1614.
- Veettil, M.; Bandyopadhyay, C.; Dutta, D.; Chandran, B. Interaction of KSHV with Host Cell Surface Receptors and Cell Entry. Viruses 2014, 6, 4024–4046.
- Suzuki, T.; Orba, Y.; Makino, Y.; Okada, Y.; Sunden, Y.; Hasegawa, H.; Hall, W.W.; Sawa, H. Viroporin activity of the JC polyomavirus is regulated by interactions with the adaptor protein complex 3. Proc. Natl. Acad. Sci. USA 2013, 110, 18668–18673.
- Husain, M.; Moss, B. Evidence against an essential role of COPII-mediated cargo transport to the endoplasmic reticulum-Golgi intermediate compartment in the formation of the primary membrane of vaccinia virus. J. Virol. 2003, 77, 11754–11766.
- Ward, B.M.; Moss, B. Golgi network targeting and plasma membrane internalization signals in vaccinia virus B5R envelope protein. J. Virol. 2000, 74, 3771–3780.
- Wang, P.G.; Tang, D.J.; Hua, Z.; Wang, Z.; An, J. Sunitinib reduces the infection of SARS-CoV, MERS-CoV and SARS-CoV-2 partially by inhibiting AP2M1 phosphorylation. Cell Discov. 2020, 6, 71.
- Cantuti-Castelvetri, L.; Ojha, R.; Pedro, L.D.; Djannatian, M.; Franz, J.; Kuivanen, S.; van der Meer, F.; Kallio, K.; Kaya, T.; Anastasina, M.; et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 2020, 370, 856–860.
- Eifart, P.; Ludwig, K.; Bottcher, C.; de Haan, C.A.; Rottier, P.J.; Korte, T.; Herrmann, A. Role of endocytosis and low pH in murine hepatitis virus strain A59 cell entry. J. Virol. 2007, 81, 10758–10768.
- Pu, Y.; Zhang, X. Mouse hepatitis virus type 2 enters cells through a clathrin-mediated endocytic pathway independent of Eps15. J. Virol. 2008, 82, 8112–8123.
- Ang, F.; Wong, A.P.; Ng, M.M.; Chu, J.J. Small interference RNA profiling reveals the essential role of human membrane trafficking genes in mediating the infectious entry of dengue virus. Virol. J. 2010, 7, 24.
- Yasamut, U.; Tongmuang, N.; Yenchitsomanus, P.T.; Junking, M.; Noisakran, S.; Puttikhunt, C.; Chu, J.J.; Limjindaporn, T. Adaptor Protein 1A Facilitates Dengue Virus Replication. PLoS ONE 2015, 10, e0130065.
- Ricotta, D.; Conner, S.D.; Schmid, S.L.; von Figura, K.; Honing, S. Phosphorylation of the AP2 mu subunit by AAK1 mediates high affinity binding to membrane protein sorting signals. J. Cell Biol. 2002, 156, 791–795.
- Jackson, L.P.; Kelly, B.T.; McCoy, A.J.; Gaffry, T.; James, L.C.; Collins, B.M.; Honing, S.; Evans, P.R.; Owen, D.J. A large-scale conformational change couples membrane recruitment to cargo binding in the AP2 clathrin adaptor complex. Cell 2010, 141, 1220–1229.
- Xiao, F.; Wang, S.; Barouch-Bentov, R.; Neveu, G.; Pu, S.; Beer, M.; Schor, S.; Kumar, S.; Nicolaescu, V.; Lindenbach, B.D.; et al. Interactions between the Hepatitis C Virus Nonstructural 2 Protein and Host Adaptor Proteins 1 and 4 Orchestrate Virus Release. mBio 2018, 9, e02233-17.
- Li, X.; Niu, Y.; Cheng, M.; Chi, X.; Liu, X.; Yang, W. AP1S3 is required for hepatitis C virus infection by stabilizing E2 protein. Antiviral Res. 2016, 131, 26–34.
- Neveu, G.; Barouch-Bentov, R.; Ziv-Av, A.; Gerber, D.; Jacob, Y.; Einav, S. Identification and targeting of an interaction between a tyrosine motif within hepatitis C virus core protein and AP2M1 essential for viral assembly. PLoS Pathog. 2012, 8, e1002845.
- Snyers, L.; Zwickl, H.; Blaas, D. Human rhinovirus type 2 is internalized by clathrin-mediated endocytosis. J. Virol. 2003, 77, 5360–5369.
- Bhattacharyya, S.; Hope, T.J.; Young, J.A. Differential requirements for clathrin endocytic pathway components in cellular entry by Ebola and Marburg glycoprotein pseudovirions. Virology 2011, 419, 1–9.
- Bekerman, E.; Neveu, G.; Shulla, A.; Brannan, J.; Pu, S.Y.; Wang, S.; Xiao, F.; Barouch-Bentov, R.; Bakken, R.R.; Mateo, R.; et al. Anticancer kinase inhibitors impair intracellular viral trafficking and exert broad-spectrum antiviral effects. J. Clin. Investig. 2017, 127, 1338–1352.
- Garrison, A.R.; Radoshitzky, S.R.; Kota, K.P.; Pegoraro, G.; Ruthel, G.; Kuhn, J.H.; Altamura, L.A.; Kwilas, S.A.; Bavari, S.; Haucke, V.; et al. Crimean-Congo hemorrhagic fever virus utilizes a clathrin- and early endosome-dependent entry pathway. Virology 2013, 444, 45–54.
- Keren, T.; Roth, M.G.; Henis, Y.I. Internalization-competent influenza hemagglutinin mutants form complexes with clathrin-deficient multivalent AP-2 oligomers in live cells. J. Biol. Chem. 2001, 276, 28356–28363.
- Wang, G.; Jiang, L.; Wang, J.; Zhang, J.; Kong, F.; Li, Q.; Yan, Y.; Huang, S.; Zhao, Y.; Liang, L.; et al. The G Protein-Coupled Receptor FFAR2 Promotes Internalization during Influenza A Virus Entry. J. Virol. 2020, 94, e01707-19.
- Sun, W.; McCrory, T.S.; Khaw, W.Y.; Petzing, S.; Myers, T.; Schmitt, A.P. Matrix proteins of Nipah and Hendra viruses interact with beta subunits of AP-3 complexes. J. Virol. 2014, 88, 13099–13110.
- Mattera, R.; Farias, G.G.; Mardones, G.A.; Bonifacino, J.S. Co-assembly of viral envelope glycoproteins regulates their polarized sorting in neurons. PLoS Pathog. 2014, 10, e1004107.
- Kolokoltsov, A.A.; Deniger, D.; Fleming, E.H.; Roberts, N.J., Jr.; Karpilow, J.M.; Davey, R.A. Small interfering RNA profiling reveals key role of clathrin-mediated endocytosis and early endosome formation for infection by respiratory syncytial virus. J. Virol. 2007, 81, 7786–7800.
- Krzyzaniak, M.A.; Zumstein, M.T.; Gerez, J.A.; Picotti, P.; Helenius, A. Host cell entry of respiratory syncytial virus involves macropinocytosis followed by proteolytic activation of the F protein. PLoS Pathog. 2013, 9, e1003309.
- Ward, C.; Maselko, M.; Lupfer, C.; Prescott, M.; Pastey, M.K. Interaction of the Human Respiratory Syncytial Virus matrix protein with cellular adaptor protein complex 3 plays a critical role in trafficking. PLoS ONE 2017, 12, e0184629.
- Nishimura, N.; Plutner, H.; Hahn, K.; Balch, W.E. The delta subunit of AP-3 is required for efficient transport of VSV-G from the trans-Golgi network to the cell surface. Proc. Natl. Acad. Sci. USA 2002, 99, 6755–6760.
- Cureton, D.K.; Massol, R.H.; Saffarian, S.; Kirchhausen, T.L.; Whelan, S.P. Vesicular stomatitis virus enters cells through vesicles incompletely coated with clathrin that depend upon actin for internalization. PLoS Pathog. 2009, 5, e1000394.
- Fujisawa, R.; Masuda, M. Ecotropic murine leukemia virus envelope protein affects interaction of cationic amino acid transporter 1 with clathrin adaptor protein complexes, leading to receptor downregulation. Virology 2007, 368, 342–350.
- Buffalo, C.Z.; Iwamoto, Y.; Hurley, J.H.; Ren, X. How HIV Nef Proteins Hijack Membrane Traffic To Promote Infection. J. Virol. 2019, 93, e01322-19.
- Pereira, E.A.; daSilva, L.L. HIV-1 Nef: Taking Control of Protein Trafficking. Traffic 2016, 17, 976–996.
- Jia, X.; Weber, E.; Tokarev, A.; Lewinski, M.; Rizk, M.; Suarez, M.; Guatelli, J.; Xiong, Y. Structural basis of HIV-1 Vpu-mediated BST2 antagonism via hijacking of the clathrin adaptor protein complex 1. Elife 2014, 3, e02362.
- Chu, H.; Wang, J.J.; Spearman, P. Human immunodeficiency virus type-1 gag and host vesicular trafficking pathways. Curr. Top. Microbiol. Immunol. 2009, 339, 67–84.
- Alford, J.E.; Marongiu, M.; Watkins, G.L.; Anderson, E.C. Human Immunodeficiency Virus Type 2 (HIV-2) Gag Is Trafficked in an AP-3 and AP-5 Dependent Manner. PLoS ONE 2016, 11, e0158941.
- Hirao, K.; Andrews, S.; Kuroki, K.; Kusaka, H.; Tadokoro, T.; Kita, S.; Ose, T.; Rowland-Jones, S.L.; Maenaka, K. Structure of HIV-2 Nef Reveals Features Distinct from HIV-1 Involved in Immune Regulation. iScience 2020, 23, 100758.
- Noble, B.; Abada, P.; Nunez-Iglesias, J.; Cannon, P.M. Recruitment of the adaptor protein 2 complex by the human immunodeficiency virus type 2 envelope protein is necessary for high levels of virus release. J. Virol. 2006, 80, 2924–2932.
- Cooper, A.; Shaul, Y. Clathrin-mediated endocytosis and lysosomal cleavage of hepatitis B virus capsid-like core particles. J. Biol. Chem. 2006, 281, 16563–16569.
- Huang, H.C.; Chen, C.C.; Chang, W.C.; Tao, M.H.; Huang, C. Entry of hepatitis B virus into immortalized human primary hepatocytes by clathrin-dependent endocytosis. J. Virol. 2012, 86, 9443–9453.
- Iwamoto, M.; Saso, W.; Nishioka, K.; Ohashi, H.; Sugiyama, R.; Ryo, A.; Ohki, M.; Yun, J.H.; Park, S.Y.; Ohshima, T.; et al. The machinery for endocytosis of epidermal growth factor receptor coordinates the transport of incoming hepatitis B virus to the endosomal network. J. Biol. Chem. 2020, 295, 800–807.




