
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Pius N. Nde | + 5402 word(s) | 5402 | 2021-03-31 06:01:37 | | | |
| 2 | Peter Tang | Meta information modification | 5402 | 2021-06-11 03:44:07 | | |
Video Upload Options
A set of key regulators whose function still needs to be fully elucidated are small non-coding RNAs (sncRNAs). Due to their broad range of unfolding functions in the regulation of gene expression during transcription and translation, sncRNAs are becoming vital to many cellular processes. Within the past decade, a novel class of sncRNAs called PIWI-interacting RNAs (piRNAs) have been implicated in various diseases, and understanding their complete function is of vital importance. Compounding evidence suggests that piRNAs encompass a wider functional range than small interfering RNAs (siRNAs) and microRNAs (miRNAs), which have been studied more in terms of cellular homeostasis and disease.
1. Introduction
Understanding the interplay between noncoding and coding RNAs represents a fast-emerging field in biomedical research. Noncoding RNAs are classified as either long non-coding RNAs (lncRNAs), which are usually longer than 200 nucleotides (nt) and small non-coding RNAs (sncRNAs) that are anywhere between 18–35 nt in length. The sncRNA that are currently known are classified into three main groups: small interfering RNAs (siRNAs, about 21 nt), microRNAs (miRNAs, about 22 nt), and Piwi-interacting RNAs (piRNAs, 24–32 nt in length) [1][2][3]. Although siRNAs and miRNAs have been studied more extensively, piRNA biogenesis and function has not been investigated in many species [4][5][6][7][8][9][10][11][12]. However, most of the knowledge about piRNA the biogenesis and function come from studies in Drosophila. Currently, piRNAs are thought to be the most abundant sncRNA within the genome, with over 30,000 members, excluding growing numbers of piRNA isoforms recently discovered across other species [13][14]. Characterization of piRNAs is dependent upon the binding with PIWI subfamily of Argonaute AGO/PIWI proteins. Argonaute family proteins are RNA binding proteins containing the highly conserved Piwi-Argonaute-Zwille domain (PAZ), the C-terminal PIWI domains, the less conserved N-terminal (N) and Middle (Mid) domains [15]. The PAZ domain functions as an RNA binding motif, and the PIWI domain retains RNase H fold domain, homologous to RNase H, an endonuclease [16]. Thus, Argonaute proteins are divided into two categories: AGO subfamily and PIWI subfamily. The PIWI subfamily proteins have been shown to be expressed within the germline and adult stem cells, while the AGO subfamily of proteins is ubiquitously expressed in most tissues. However, the literature shows that PIWI subfamily of proteins are also expressed in somatic cells, suggesting that piRNAs may exhibit roles outside of germline integrity maintenance [17][18]. PIWI family proteins have also been identified in mice and humans as MIWI and HIWI subfamilies, respectively [19][20]. The piRNA/Piwi complex interacts with various proteins that facilitate many functions, including: piRNA biogenesis, transposon silencing, chromatin remodeling, transcriptional and translational regulation [21]. Within the past decade, increasing number of reports implicate piRNAs as modulators of various diseases including cardiomyopathies, cancer, and infectious diseases.
2. Structure and Function of piRNAs
2.1. piRNA Structure and Biogenesis
P-element induced wimpy testis interacting RNAs (piRNAs) are the most recently characterized class of small noncoding RNAs (sncRNAs). These RNA molecules are found in germline and somatic cells. Their sizes range anywhere from 24–31 nucleotides long and many reports identify a 5′ terminal uridine or tenth adenosine [22]. These sncRNAs have a unique feature in which a 2′ O-methyl group on the 3′ ribose is required for maturation [23][24]. This 2′ O-methyl group addition is performed by the piRNA methyltransferase Hen1 (HENMT1), a step that is conserved in all organisms [24]. Loss of Henmt1 caused piRNA instability via decreased piRNA abundance and size, as illustrated by Lim et al. [25]. Mature piRNAs interact with a specific subset of argonaute proteins called p -element induced wimpy testis (PIWI) proteins; these RNA binding proteins interact with piRNAs, forming a ribonucleoprotein complex known as the piRNA induced silencing complex (piRISC). The piRISC is guided by piRNAs to recognize sequences at the chromatin, transcriptional, and post-transcriptional levels. Proteins containing Tudor domains (TDRDs) also associate with PIWI proteins and have been shown to play an active role in the piRNA metabolic pathway. TDRDs expedite piRISC action as well as bolster the stability of PIWI proteins by acting as a molecular scaffold [26].
Biogenesis of sncRNA generally occurs in a Dicer/Drosha dependent manner, in which double-stranded RNA precursor molecules are remodeled into functional sncRNAs [27]. Alternatively, piRNAs participate in a distinct process known as primary and secondary piRNA biogenesis [21]. This process, which has been partially described and characterized in Drosophila, is also conserved in C. elegans. The long, single-stranded piRNA precursor molecules are derived from the transcripts of protein coding genes, transposons, tRNA, rRNA, and intergenic loci [28][29][30][31] which possibly contributes to the targeted sequence specificity observed in mature piRNAs. piRNAs mostly originate from repetitive sequences within the genome including transposable elements (TEs), which can be distributed in clusters throughout the chromosomes on both DNA strands [4].
Primary piRNA biogenesis begins with transcription of single stranded piRNA progenitor molecules by RNA polymerase II, albeit recruitment mechanisms and promoter regions remain elusive [32] constituting new areas of research. Processing of these precursors operates in a Dicer/Drosha independent manner. The single stranded piRNA precursor molecules are shuttled out of the nucleus into Yb bodies, cytoplasmic membranesless organelles [33] (Figure 1). In the Yb bodies, the precursor piRNA/Piwi complex interacts with TDRD, Armitage, Vreteno, and Sister of Yb proteins. The function of these proteins during piRNA biogenesis remains elusive, however, they have been shown tso colocalize to Yb bodies in Drosophila germline, suggesting that they play instrumental role in piRNA biogenesis [34][35][36][37][38][39]. Yb bodies have been shown to be surrounded by mitochondria, facilitating the mitochondria membrane anchored endonuclease Zucchini to process precursor piRNAs into smaller segments [35][40]. With the help of Shutdown (Shu) and Heat shock protein 90 (Hsp90), piRNAs then complex with Piwi proteins [41][42]. Lastly, the 3′ end is processed by an unknown protein, after which the HENMT1 adds a methyl group to the 2′ carbon of the ribose on the 3′ end of the transcript [24][43]. This modification is crucial for piRNA stability [25]. Mature piRNA complexes can either reenter the nucleus to induce transposon silencing and gene regulation or initiate secondary piRNA biogenesis, also known as the ping pong cycle [4][44]. Interestingly, different piRNA subsets have been shown to complex with different PIWI proteins. Piwi and Aubergine (Aub) have a propensity for piRNA sequences that are specifically antisense to transposons with a 5′ uridine. In contrast, Ago3 proteins bind to sense piRNAs containing adenine at the 10th nt position, with no preference for 5′ U piRNA sequences [4][7][28][45] (Figure 1).
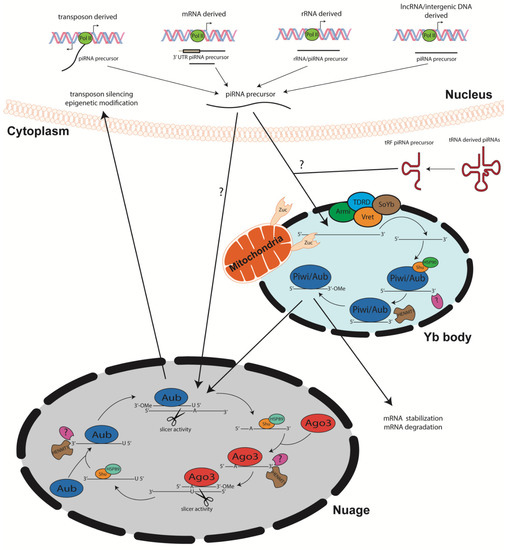
The ping-pong cycle which has been the major contributor of piRNA biogenesis, depends on Aub and Ago3 associated piRNAs. Once long, single stranded precursor piRNAs are transcribed via RNA polymerase II, they are shuttled to the nuage, an electron-dense phase separated granules anchored to the cytoplasmic face of the nuclear pore [44] TDRD proteins may serve as scaffolds and initiators of ping pong cycle, as mutations within these proteins disrupt piRNA biogenesis [40][46][47][48]. The amplification cycle involving these PIWI proteins calls for mature 5′ U piRNAs complexed with Aub to recognize specific complementary sequences within the transcript. Once complementarity between the U:A nt is achieved, the Aub enacts endonucleolytic cleavage of the target piRNA at the 10th/11th nt of the sequence. This slicing activity generates the 5′ end of a new sense piRNA, including a 10 nt long 5′ overlap with the initial antisense piRNA and an adenosine residue at the 10th nt. Shu and Hsp89 are hypothesized to load the newly generated piRNA onto Ago3 [41][42]. Subsequent trimming by an unidentified enzyme and 3′ modification by piRNA methyltransferase finalizes piRNA maturation. The newly formed complex then produces more antisense Aub-bound piRNAs from piRNA clusters within the transcripts via the same biogenesis mechanism. The secondary piRNA amplification of the ping pong cycle has been linked to targeted post-transcriptional gene silencing [4][45]. piRNA biogenesis from sense/antisense transposons has been defined but requires further investigation [49]. However, others have shown that piRNAs derived from both mRNA and lncRNA are typically generated from the 3′ UTR but the mechanism is yet to be fully unraveled. Noncanonical by-products of concurrent transcription of adjacent genes are also thought to be piRNA precursor transcripts, which hail from dual-stranded clusters [50]. Though much of the piRNA biogenesis pathway is conserved amongst most organisms [51], further studies are needed to fully elucidate mechanisms within the pathway (Figure 1).
2.2. Transposon Silencing
One of the earliest functions of piRNAs is their ability to serve as regulators of TE movement throughout the genome. TEs can be randomly inserted throughout the genome, causing changes in protein coding genes and regulatory sequences, which can affect gene expression and cause the production of defective or malfunctioning proteins [52]. It is well documented that piRNAs are vital to ensuring genomic integrity, as the piRNAs/Piwi complex can monitor TE activity by silencing TEs post transcriptionally [53]. Mutations within the germline have been shown to cause an increase in retrotransposons, which causes the death of germ cells in conjunction with various deficiencies within microtubule cytoskeletal polarization. Consequently, alterations in this conserved mechanism leads to changes in polarized localization of specific proteins and mRNAs that are necessary for oogenesis in Drosophila [54].
Sarot et al. illustrated that Piwi is required for silencing transposons within the 3am locus in gonadal somatic cells, which is a well-known active region [55]. A vast number of piRNAs originate from transposon regions of the genome, as shown by others and recently our group [56][49][57][58]. Several studies have demonstrated the importance of PIWI proteins in the regulation of transposon silencing in Drosophila. For example, aub-mutants illustrated increased transposon activity [54][59][60][61][62]. Similarly, Ago3 and Aub are known to colocalize in the nuage for secondary piRNA biogenesis; mutations within either of these proteins triggered defects in biogenesis and elevated transposon transcripts [63]. The vital function of piRNAs in maintaining genome stability has been illustrated using knockout models designed to understand the roles of PIWI proteins [8][64]. In germ cells of mili/miwi2 KO mice, Carmell et al. observed a significant increase in transposable elements [64]. Transposon silencing remains a critical function of piRNAs, however further studies should aim to describe the targeted action of piRNAs as well uncover other mechanisms that can contribute to this silencing.
2.3. Epigenetic Regulation
piRNAs have been shown to participate in epigenetic regulation through two mechanisms: activation of sequence specific de novo methylation and chromatin remodeling. Huang et al. demonstrated a vital role of the piRNA/Piwi interface in epigenetic programing in Drosophila. Once piRNA complementary sequences were introduced in ectopic sites, several DNA binding proteins, including Piwi and heterochromatin protein 1 (HP1a), induced increased H3K9me2/3 recruitment, which ultimately led to reduced RNA polymerase II enlistment. This finding supports the hypothesis that piRNAs are necessary for the recruitment of epigenetic associated proteins to precise genomic regions/locations in Drosophila [65][66]. Deletion of PIWI proteins in murine models significantly altered the epigenetic makeup throughout the genome [8][64]. Watanabe et al. demonstrated simultaneous coactivation of piRNA-mediated DNA methylation and transcription of piRNA dependent regions in mice. Retrotransposons in piRNA dependent regions and in piRNA clusters were knocked out in a mouse model. The deleted regions were shown to play a role in the initiation of piRNA-mediated methylation. These results showed the importance of MIWI in determining chromatin structure through direct targeting of piRNAs to genomic regions, which also has been observed in other organisms [67].
The synergic interactions between methyltransferases, histone modifying proteins, chromatin remodeling proteins, and piRNA/Piwi complex is implicated in de novo methylation and subsequent suppression of TEs within the germline. However, the mechanism is not well understood and necessitates further investigation. In Drosophila, Piwi has been shown to interact with chromosomes in somatic tissues, implicating its role in inducing epigenetic modifications at various binding sites [66][68][69]. A study showed that a piRNA/Piwi complex in Aplysia nerve cells promoted methylation of CpG islands within the promoter region of transcription factor CREB2, further solidifying the role of piRNA/PIWI complex in epigenetic gene regulation [70][71]. piRNA/Piwi complexes can also recruit DNA methyltransferases to target CpG sites in non-TE protein coding regions, which alters transcriptional activity in mice [72]. Fu et al. demonstrated that genomic regions neighboring differentially methylated CpG areas were fortified with sequence matches to transfected piRNAs in somatic cancer cell lines [73]. This transcriptional repression can then possibly be inherited at the target sites in mammals, however further research is needed to elucidate this fully. For example, overexpression of piR-021285 induced methylation of ARHGAP11A at CpG site within the 5′ UTR/first exon, which dampened messenger RNA expression while simultaneously inhibiting breast cancer programmed cell death [74]. Recent efforts to uncover mechanistic machinery of piRNA directed DNA methylation in mammals have identified proteins essential to this function. SPOCD1 has recently been identified as a MIWI2 associated protein that is required for TE silencing via piRNA mediated methylation. Loss of Spocd1 induced infertility in male mice without altering piRNA biogenesis or MIWI2 nuclear localization [75]. Similarly, recruitment of TEX15 has been shown to be an executor of piRNA directed methylation, as TEX15 is required for the nuclear function of MIWI2 [76]. Further research should delineate exact mechanisms that trigger and induce piRNA/Piwi dependent methylation throughout the genome. Inheritance of these piRNA induced epigenetic modifications should also be explored in depth.
Although the mechanism of piRNAs induce methylation of genomic regions is unclear, studies have provided evidence indicating that piRNAs serve as guides for histone modifying and chromatin remodeling proteins. Mohn et al. utilized chromatin immunoprecipitation to suggest that the ribonucleoprotein complex targets sequences within euchromatin via preliminary transcripts, while also targeting heterochromatin via RNA/DNA interaction. This is a direct result of piRNA sequence complementarity, illustrating the wide yet potentially specific target [18][77]. Piwi has been found to interact with HP1a directly, and colocalization of these proteins has been observed on many sites throughout chromosomes in Drosophila [65][66]. As regulators of the position effect variegation (PEV), which decreases gene expression of euchromatin genes near heterochromatin sites, Piwi and Aub can silence gene expression via heterochromatin assembly [68]. piRNA dependent recruitment of histone modifying proteins remains a mystery, however current research should continue to decode this mechanism.
2.4. Post-transcriptional and Translational Control
In several organisms, it has been shown that 3′UTRs of protein coding mRNAs are sources for piRNA production in Drosophila, mice, and Xenopus, indicating that piRNAs also can regulate mRNA turnover [78]. It was observed that VAS protein levels increased in aub and ago3 mutants, which could possibly be attributed to decreased number of piRNAs derived from vas mRNA [63][79]. Miwi proteins in mice have been shown to complex with and stabilize mRNAs of genes necessary for post-meiotic steps of spermatogenesis [80]. Lee et al. showed that piRNAs could target non-transposon genes that contribute to the regulation of spinal shape [81].
In terms of piRNA/mRNA interaction, piRNAs require base pairing at both the 2–11 nt at the 5′ end of the piRNA as well as within the 12–21 nt of the sequence [74]. To regulate mTOR expression, piR-55490 was observed to bind to the 3′ UTR, initiating mRNA degradation and muffle lung cancer development [82]. Esposito et al. concluded that testis and brain specific piRNAs can regulate MTNR1A expression in human somatic cells [83]. piRNAs have also been shown to play a role in mRNA decay via deadenylation of the 3′ end of mRNA transcripts. In Drosophila, piRNA dependent degradation of maternal mRNA is dependent upon Smg, deadenylase CCR4, and the piRNA/Piwi complex. Impaired piRNA regulation induced transcript stabilization [84][85]. The post-transcriptional/translational regulatory role of piRNAs outside of the germ line has yet to be fully understood, however these studies strongly implicate piRNAs as critical post-transcriptional regulators.
Evidence of chromatin remodeling, de novo methylation, and direct transcriptional regulation shows the multifaceted roles of piRNAs [74][56][52][53][54][55][57][58][59][60][61][62][63][64][65][66][67][68][70][71][72][73][75][76][77]. These processes clearly classify piRNAs as regulators of gene expression in various capacities. Currently, little evidence implicates the piRNA/Piwi complex in translational regulation, however several emerging reports indicate that PIWI proteins can interact with translational proteins [86][87][88]. For example, Grivna et al. showed that Miwi binds to eIF4E, the mRNA 5′ cap binding protein essential for guidance to ribosomes in the cytoplasm and translational control [86]. A separate study reported that Mili/eIF3a complex interacts with eIF4E/eIF4G 5′ cap binding complex [88]. Recently, Ramat et al. reported that PIWI protein Aub is required for translational activation of nanos mRNA, which is essential in germ line survival and pluripotency. Through direct physical interaction, Aub complexes with poly(A)-binding protein (PABP) and translation initiation factor eIF3. Using polysome gradient profiling, Aub was shown to be essential for the initiation step of translation [87]. These studies suggest PIWI proteins potentially play a role in initiation and activation of translation; however, further studies are needed to completely delineate the functions of PIWI proteins and piRNAs in this process.
3. piRNAs vs other sncRNAs
Although piRNAs are the most newly identified class of sncRNAs, they are the least classified. Hence, several research efforts have been geared towards delineating similarities and differences between piRNAs and other sncRNAs, specifically miRNAs and siRNAs. piRNAs are widely and differentially expressed in various tissues and cell types, but mostly have been detected within the germ cells of mammals, fish, and Drosophila [89][90]. In terms of biogenesis, it is well understood that piRNAs diverge significantly from miRNAs and siRNAs. piRNAs are generated via RNaseIII-independent pathways and do not involve dsRNA progenitors. Since these unique sncRNAs are generated from long single stranded precursors, they show preferential cleavage at uridine residues and then complex with PIWI proteins [49]. In contrast, siRNAs are processed by Dicer from small dsRNA complexes with a distinct 2 nt 3′ overhang and 5′ phosphate group. miRNAs are transcribed via RNA polymerase II from primary miRNA (pri-miRNA) precursors which contain stem loop structure. These molecules undergo further processing that give rise to classical miRNAs [91][92].
Structurally, miRNAs are short, single stranded noncoding RNAs, generally 20–24 nucleotides in length, which are encoded in eukaryotic cells and function in various signaling pathways [93][94]. siRNAs, however, are 21–23 nt long RNA duplexes that interact and activate the RNA-induced silencing complex (RISC) [95][96]. Functionally, miRNAs are recognized for control of mRNA stability and translation. These sncRNAs are found within the cytoplasm where they recognize and bind the 3′ UTR of target mRNA complementary sequences via the miRNA “seed sequence” located at their 5′ end [97]. The consequence of this interaction is either mRNA degradation or translational repression. In contrast, siRNAs must accomplish 100% complementarity with mRNA target seqence in order to enact repressive function [96].Though piRNAs and miRNAs can bind to various target molecules, siRNAs are specific to one mRNA target and regulate expression exclusively through endonucleolytic cleavage via RISC [96]. miRNAs are categorized via sequence conservation and the seed sequence, meaning many miRNAs within the same family share similar targets in intertwining signaling pathways [98]. Current research reveals untraditional roles of miRNAs within the nucleus, including transcriptional silencing/activation, and alternative gene splicing [99][100][101]. We also recently showed the role of miRNA in translation initiation [102]. Although the mechanisms which activate these noncanonical roles remain to be fully understood, several debates regarding miRNA nuclear translocation persist [101]. Several studies have reported that miRNAs activate gene expression via promoter region binding by utilizing sequence complementarity [99][103].
In contrast, piRNAs are about 24-31 nucleotides long and were discovered in Drosophila mutants that underwent unequal division of stem cells within the germline. As the most abundant sncRNAs [13][14], extensive research has begun to uncover the vast roles these molecules play in cell homeostasis and disease. Developmentally, they are essential for germ line survival and enact transposon silencing, actively maintaining genome integrity. Any defects and disruptions in piRNA biogenesis is detrimental, causing germline specific cell death and sterility via upregulation of transposon expression as seen in mice and fish [104]. Other functions include conserving telomere structure, controlling RNA silencing, and inducing epigenetic factors that alter chromatin structure. More recently, studies have begun to investigate piRNA function in somatic cells, where expression seems to be regulated in a tissue specific manner. Targeting of mRNA sequences is believed to be similar to that of miRNAs, specifically utilization of the seed sequence. Unlike miRNA, Piwi/piRNA complexes can also recruit other proteins such as CCR4-NOT and Smg to create RISC like complexes called pi-RISCS, which repress mRNA translation via imperfect base pairing [105][106]
For piRNAs to enact their gene silencing function, they must complex with a specific subset of argonaute proteins called PIWI proteins. The association with Piwi and Dicer independent mechanism are the main distinguishing factors between siRNAs and piRNAs. However, similarly to other sncRNAs, piRNAs are loaded into the PIWI protein and guide argonaute to target sequences using Watson-Crick base pairing and seed sequence complementarity. When complementarity is reached, RNase H-fold activity of the PIWI domain cleaves phosphodiester bond of two nucleotides in the target RNA that pair with the 10th and 11th nt of the piRNA sequence [49]. In contrast, Dicer within RISC displays RNaseIII activity, initiating two cuts which are 21–22 nt apart on the target sequence [107].
Post transcriptional functions of piRNA are poorly understood, and only a few reports show different PIWI proteins complexing with mRNA 5′ cap and eIF proteins to regulate translation [86][88]. Extensive research is needed in this area to fully elucidate this potential function. In contrast, miRNAs and siRNAs are known mediators of post transcriptional regulation, and the pathways they regulate differ according to cell type, organism, and argonaute protein binding. Argonaute proteins complexed with miRNAs or siRNAs utilize slicer activity and/or slicer dependent mechanisms to employ repression functionality. This includes inhibition of eIF4E cap binding and mobilization of other proteins that participate in translational repression and mRNA degradation [85][108]. High complementarity is required for target mRNA degradation via slicing, mediated by degradation proteins such as XRN1, exosomes and Ski complexes [109]. Translationally repressed RNAs can be degraded through deadenylation, decapping, and subsequent degradation in P-bodies [84][85]. Though many differences reside in biogenesis and many similarities reside in function of sncRNAs, further studies are required to identify the unique roles that each of these RNAs play in the regulation of gene expression.
4. The Role of piRNAs in Cardiomyopathies
It was previously thought that piRNAs expression was exclusive to germ cell lines, but recent studies have shown that piRNAs are also expressed in non-germline cells [56][110]. In recent times, it has been theorized that piRNAs are expressed in higher eukaryotic genomes. This theory came from the discovery that piRNAs regulate transposons and higher eukaryotes house large repositories of transposable elements. Recent advances indicate that piRNAs are found in the central nervous system, liver, cardiovascular system, and in circulating serum exosomes [111][112]. Researchers have recently taken interest in the role of piRNA expression in the differentiation of cardiomyocytes.
Greca et al. sought to investigate the role of piRNAs in the development of cardiac progenitor cells. To examine this, the group analyzed RNA-seq data from H9 human embryonic stem cells, early mesoderm progenitor cells and cardiomyocytes. Stem-loop retrotranscription primers were generated to obtain cDNA from piRNAs of interest and real time PCR was carried out to quantify piRNA expression. It should be noted that mapped reads from mRNAs and other non-coding RNAs were filtered out by size to accurately determine the role of piRNAs in the development of cardiac progenitor cells. The group reported that there are at least 447 piRNAs involved in the progression of pluripotent stem cells to cardiomyocytes. Furthermore, cardiac progenitor cells expressed increased levels of mitochondrial tRNA/rRNA derived piRNAs, followed by a significant upregulation in HIWI2 expressing cells [110].
Since the majority of these piRNAs were derived from other types of RNAs, the group noted that further validation must be performed to ensure their accurate classification. However, approximately 90 of the 447 piRNAs involved in the development of cardiomyocytes originated from the mitochondrial genome [97]. It should be noted that piRNAs were found to be uniformly scattered throughout the genome, aside from the mitochondrial genome where they were expressed in clusters. RNA-seq data suggested that nuclear encoded lncRNA MALAT1 and TTN encompass the highest number of piRNAs in cardiac progenitor cells. RNA-seq data demonstrated that MALAT1-derived piRNAs include piR-4403262, and piR-4424378. TTN-derived piRNAs include piR-1551388, piR-4193743, and piR-2715002. Interestingly, the group reported that six times more piRNAs were downregulated in cardiac progenitor cells compared to pluripotent stem cells. This study showed that piRNAs are involved in cardiac cell differentiation and maturation, thus dysregulation of piRNA can result into cardiac pathology [110].
Vella et al. also investigated the role of the piRNome, the global expression profile of piRNAs within the cellular system, in different cardiac cell types using biopsies from patients undergoing heart surgery. These biopsies were used to isolate cardiac progenitor cells, which were grown as undifferentiated self-adherent 3D cultures in suspension (cardiospheres) or as adherent monolayers on a plate (cardiosphere-derived cells). They also isolated cardiac fibroblasts from these biopsies and examined the piRNome among each of these cell types via microarray analysis and RT-PCR validation. The group reported that 15,311 piRNAs were expressed in all three cell types, among the 23,000 piRNAs within the dataset used for the study. Of the identified piRNAs, 641 were upregulated and 1,381 were downregulated in cardiospheres compared to cardiosphere-derived cells. However, there were only 255 upregulated and 780 downregulated piRNAs in cardiospheres compared to cardiac fibroblasts. Furthermore, when cardiosphere-derived cells were compared to cardiac fibroblasts, only 52 piRNAs were upregulated and 129 were downregulated. These findings suggest that piRNAs are differentially expressed in cardiac cells and have distinct functions in individual cell types. To further support this theory, the group reported that certain piRNAs such as DQ570326 and DQ58246 are expressed at higher levels in cardiosphere-derived cells compared to cardiospheres. Furthermore, some piRNAs, such as DQ579896 and DQ581624 are upregulated in cardiac fibroblast but not cardiospheres or cardiosphere-derived cells [113].
Di Giacomo et al. reported that the inhibition of LINE-1 retrotransposons was mediated by piRNAs [114], and this mechanism was shown to attenuate ischemic heart disease via piRNA-mediated activation of AKT [115]. Vella et al. examined LINE targets of upregulated piRNAs in cardiospheres, cardiosphere-derived cells and cardiac fibroblasts and they found that the upregulated piRNAs targeted all three classes of LINE reverse transcriptase: LINE-1, LINE-2 and CR1. More specifically, the transcript levels of LINE-1 transcripts were lower in cardiospheres and cardiosphere-derived cells compared to cardiac fibroblasts. This corresponded to increased expression of phosphorylated AKT in cardiospheres and cardiac derived cells compared to cardiac fibroblast. It was also demonstrated that piRNA-mediated phosphorylation of the Ser473 residue of AKT was associated with the phosphorylation of the Ser9 residue of GSK3β to inactivate GSK3β’s kinase activity in cardiospheres and cardiosphere derived cells. Hence, piRNA expression is associated with cardiac regeneration and mediated by AKT-mediated phosphorylation. However, it is possible that cardiac piRNA expression has downstream effects on cellular signaling pathways that involve GSK3β, which is a known repressor of cardiac hypertrophy by phosphorylating proteins involved in cell proliferation such as β-catenin and NFATC4 [116]. Overall, this study showed that the piRNome may play a role in regeneration within cardiac progenitor cells—indicating that piRNAs may be involved in cardiac pathogenesis and may serve as therapeutic targets [113].
In 2020, Gao et al. investigated the potential role of piRNAs in cardiac hypertrophy by examining the global expression of piRNAs in mice. Samples were obtained from the left ventricle 4 weeks after a transverse aortic construction surgery, then subjected to microarray analysis. The group found that piR-141981, piR-6999 and piR-110550 were increased but piR-13375 and piR-106654 were decreased in mice that underwent a transverse aortic construction surgery versus a placebo/sham surgery. Of these piRNAs, piR-141981, piR-110550 and piR-13375 increased mRNA expression of β-myosin heavy chain, a cardiac stress marker and biomarker of cardiac hypertrophy. The group also examined the global expression of these three piRNAs and found that piR-141981 was expressed at higher levels in the heart, compared to other organs [117].
Since piR-141981 was previously unclassified, the group named it the cardiac-hypertrophy-associated piRNA (CHAPIR). CHAPIR was found to be expressed at higher levels in cardiomyocytes compared to cardiac fibroblasts. In mice, CHAPIR is located on chromosome 4 within intron 1 of Gm12648. The group generated CHAPIR knockout mice to observe the role of this piRNA in the development of hypertrophy following traverse aortic construction surgery. They found that a deficiency of CHAPIR blocked the development of cardiac hypertrophy following surgery. This was mediated through the attenuation of hypertrophic biomarkers including the expression of Anp, Myh7 and Bnp. It should be noted that Anp and Bnp encode atrial and brain natriuretic protein, respectively. Myh7 encodes the heavy β-myosin chain [117]. The group found that CHAPIR increases the stability of Parp10 mRNA transcripts by inhibiting METTL3-mediated N6-methyladenosine methylation of Parp10 mRNA. More specifically, Parp10 gene expression is upregulated when the CHAPIR-PIWIL4 complex interacts with METTL3, blocking N6-methyladenosine (m6A) methylation of Parp10 mRNA transcripts. Under “normal” conditions, m6A destabilizes Parp10 mRNA transcripts and accelerates their degradation. However, in the presence of CHAPIR, Parp10 mRNA is translated into PARP10, which inactivates GSK3β through mono-ADP-ribosylation. Inhibition of GSK3β kinase activity then leads to the onset of hypertrophy, marked by nuclear accumulation of NFATC4. Under “normal” conditions NFATC4 is phosphorylated by activated GSK3β—promoting its nuclear export and decreased hypertrophic response. However, when GSK3β’s kinase activity is inhibited it cannot phosphorylate NFATC4, which remains in the nucleus—promoting cardiac hypertrophy. This finding supports the theory that piRNA activity may be associated with cardiomyopathies since cardiac progenitor cells do not express large amounts of piRNAs under normal conditions [117].
Furthermore, Yang et al. demonstrated that exosomal piRNAs have the potential to serve as biomarkers in patients with heart failure. The group isolated exosomes from the blood of heart failure patients and age matched controls. RNA was isolated from the exosomes and the expression of piRNAs was examined using RNA-seq. They reported that approximately 585 piRNAs were upregulated and 4,623 were downregulated in the exosomes of patients with heart failure. Furthermore, has-piR-006426 and has-piR-0200009 were among the most downregulated piRNAs derived from the serum exosomes of heart failure patients. These findings indicate that hsa-piR-006426 which is located on chromosome 17p11.2 and has-piR-0200009, which is located on chromosome 7q35 may be involved in heart failure and may serve as clinically relevant biomarkers for heart disease [118]. In a separate study, Rajan et al. discovered that PiR_2106027 was elevated in the serum of patients with Troponin-I negative myocardial infarctions and downregulated in healthy patients. Hence, PiR_2106027 may serve as a biomarker for myocardial infarctions [119]. Furthermore, recent studies suggest a correlation between piRNAs, mTOR and cardiac tissue regeneration. Specifically, piR-55490 has been shown to regulate mTOR, which consequently promotes angiogenesis and cardiac tissue regeneration. This suggest that piRNAs may regulate both cancer and cardiovascular disease/regeneration in an mTOR-dependent manner [82][120].
Overall, these studies suggest that piRNA expression differs among cardiac progenitor cells and cardiac fibroblasts, suggesting that piRNAs may play distinct roles in different types of cells. Furthermore, cardiac progenitor cells tend to express downregulated levels of piRNAs compared to other cardiac cells such as cardiac fibroblasts. However, the dysregulation of specific piRNAs, such as CHAPIR promotes the development of cardiomyopathies, including cardiac hypertrophy. Furthermore, other piRNAs such as has-piR-0200009 may serve as clinically relevant biomarkers of cardiomyopathies and heart failure. Although the exact function of piRNAs in the progression of cardiomyopathies remain to be fully elucidated, current research suggest that piRNAs may serve as therapeutic targets and clinically relevant biomarkers.
References
- Farazi, T.A.; Juranek, S.A.; Tuschl, T. The growing catalog of small RNAs and their association with distinct Argonaute/Piwi family members. Development 2008, 135, 1201–1214.
- Leung, Y.Y.; Kuksa, P.P.; Amlie-Wolf, A.; Valladares, O.; Ungar, L.H.; Kannan, S.; Gregory, B.D.; Wang, L.-S. DASHR: Database of small human noncoding RNAs. Nucleic Acids Res. 2015, 44, D216–D222.
- Fang, Z.; Du, R.; Edwards, A.; Flemington, E.K.; Zhang, K. The Sequence Structures of Human MicroRNA Molecules and Their Implications. PLoS ONE 2013, 8, e54215.
- Brennecke, J.; Aravin, A.A.; Stark, A.; Dus, M.; Kellis, M.; Sachidanandam, R.; Hannon, G.J. Discrete Small RNA-Generating Loci as Master Regulators of Transposon Activity in Drosophila. Cell 2007, 128, 1089–1103.
- Murchison, E.P.; Kheradpour, P.; Sachidanandam, R.; Smith, C.; Hodges, E.; Xuan, Z.; Kellis, M.; Grutzner, F.; Stärk, A.; Hannon, G.J. Conservation of small RNA pathways in platypus. Genome Res. 2008, 18, 995–1004.
- Juliano, C.E.; Reich, A.; Liu, N.; Götzfried, J.; Zhong, M.; Uman, S.; Reenan, R.A.; Wessel, G.M.; Steele, R.E.; Lin, H. PIWI proteins and PIWI-interacting RNAs function in Hydra somatic stem cells. Proc. Natl. Acad. Sci. USA 2013, 111, 337–342.
- Vagin, V.V.; Sigova, A.A.; Li, C.; Seitz, H.; Gvozdev, V.; Zamore, P.D. A Distinct Small RNA Pathway Silences Selfish Genetic Elements in the Germline. Science 2006, 313, 320–324.
- Aravin, A.A.; Sachidanandam, R.; Girard, A.; Fejes-Toth, K.; Hannon, G.J. Developmentally Regulated piRNA Clusters Implicate MILI in Transposon Control. Science 2007, 316, 744–747.
- Aravin, A.A.; Sachidanandam, R.; Bourc’His, D.; Schaefer, C.; Pezic, D.; Toth, K.F.; Bestor, T.; Hannon, G.J. A piRNA Pathway Primed by Individual Transposons Is Linked to De Novo DNA Methylation in Mice. Mol. Cell 2008, 31, 785–799.
- Roovers, E.F.; Rosenkranz, D.; Mahdipour, M.; Han, C.-T.; He, N.; Lopes, S.M.C.D.S.; Van der Westerlaken, L.A.; Zischler, H.; Butter, F.; Roelen, B.A.; et al. Piwi Proteins and piRNAs in Mammalian Oocytes and Early Embryos. Cell Rep. 2015, 10, 2069–2082.
- Praher, D.; Zimmermann, B.; Genikhovich, G.; Columbus-Shenkar, Y.; Modepalli, V.; Aharoni, R.; Moran, Y.; Technau, U. Characterization of the piRNA pathway during development of the sea anemone Nematostella vectensis. RNA Biol. 2017, 14, 1727–1741.
- Gainetdinov, I.; Skvortsova, Y.; Kondratieva, S.; Funikov, S.; Azhikina, T. Two modes of targeting transposable elements by piRNA pathway in human testis. RNA 2017, 23, 1614–1625.
- Peng, J.C.; Lin, H. Beyond transposons: The epigenetic and somatic functions of the Piwi-piRNA mechanism. Curr. Opin. Cell Biol. 2013, 25, 190–194.
- Kozomara, A.; Griffiths-Jones, S. miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014, 42, D68–D73.
- Song, J.-J.; Liu, J.; Tolia, N.H.; Schneiderman, J.; Smith, S.K.; Martienssen, R.A.; Hannon, G.J.; Joshua-Tor, L. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat. Struct. Mol. Biol. 2003, 10, 1026–1032.
- Parker, J.S.; Roe, S.M.; Barford, D. Crystal structure of a PIWI protein suggests mechanisms for siRNA recognition and slicer activity. EMBO J. 2004, 23, 4727–4737.
- Cheng, E.-C.; Kang, D.; Wang, Z.; Lin, H. PIWI Proteins Are Dispensable for Mouse Somatic Development and Reprogramming of Fibroblasts into Pluripotent Stem Cells. PLoS ONE 2014, 9, e97821.
- Ross, R.J.; Weiner, M.M.; Lin, H. PIWI proteins and PIWI-interacting RNAs in the soma. Nat. Cell Biol. 2014, 505, 353–359.
- Qiao, D.; Zeeman, A.-M.; Deng, W.; Looijenga, L.H.J.; Lin, H. Molecular characterization of hiwi, a human member of the piwi gene family whose overexpression is correlated to seminomas. Oncogene 2002, 21, 3988–3999.
- Kuramochi-Miyagawa, S.; Kimura, T.; Yomogida, K.; Kuroiwa, A.; Tadokoro, Y.; Fujita, Y.; Sato, M.; Matsuda, Y.; Nakano, T. Two mouse piwi-related genes: Miwi and mili. Mech. Dev. 2001, 108, 121–133.
- Mani, S.R.; Juliano, C.E. Untangling the web: The diverse functions of the PIWI/piRNA pathway. Mol. Reprod. Dev. 2013, 80, 632–664.
- Liu, P.; Dong, Y.; Xiao-Guang, C.; Puthiyakunnon, S.; Wu, Y.; Chen, X.-G. Developmental piRNA profiles of the invasive vector mosquito Aedes albopictus. Parasites Vectors 2016, 9, 1–15.
- Kirino, Y.; Mourelatos, Z. Mouse Piwi-interacting RNAs are 2’-O-methylated at their 3’ termini. Nat. Struct. Mol. Biol. 2007, 14, 347–348.
- Saito, K.; Sakaguchi, Y.; Suzuki, T.; Siomi, H.; Siomi, M.C. Pimet, the Drosophila homolog of HEN1, mediates 2’-O-methylation of Piwi-interacting RNAs at their 3’ ends. Genes Dev. 2007, 21, 1603–1608.
- Lim, S.L.; Qu, Z.P.; Kortschak, D.R.; Lawrence, D.M.; Geoghegan, J.; Hempfling, A.L.; Bergmann, M.; Goodnow, C.C.; Ormandy, C.J.; Wong, L.; et al. HENMT1 and piRNA Stability Are Required for Adult Male Germ Cell Transposon Repression and to Define the Spermatogenic Program in the Mouse. PLoS Genet. 2015, 11, e1005620.
- Weick, E.-M.; Miska, E.A. piRNAs: From biogenesis to function. Development 2014, 141, 3458–3471.
- Welker, N.C.; Pavelec, D.M.; Nix, D.A.; Duchaine, T.F.; Kennedy, S.; Bass, B.L. Dicer’s helicase domain is required for accumulation of some, but not all, C. elegans endogenous siRNAs. RNA 2010, 16, 893–903.
- Saito, K.; Nishida, K.M.; Mori, T.; Kawamura, Y.; Miyoshi, K.; Nagami, T.; Siomi, H.; Siomi, M.C. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev. 2006, 20, 2214–2222.
- Aravin, A.; Gaidatzis, D.; Pfeffer, S.; Lagos-Quintana, M.; Landgraf, P.; Iovino, N.; Morris, P.; Brownstein, M.J.; Kuramochi-Miyagawa, S.; Nakano, T.; et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nat. Cell Biol. 2006, 442, 203–207.
- Girard, A.; Sachidanandam, R.; Hannon, G.J.; Carmell, M.A. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nat. Cell Biol. 2006, 442, 199–202.
- Grivna, S.T.; Beyret, E.; Wang, Z.; Lin, H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006, 20, 1709–1714.
- Gu, W.; Lee, H.-C.; Chaves, D.A.; Youngman, E.M.; Pazour, G.J.; Conte, D.; Mello, C.C. CapSeq and CIP-TAP Identify Pol II Start Sites and Reveal Capped Small RNAs as C. elegans piRNA Precursors. Cell 2012, 151, 1488–1500.
- Hirakata, S.; Ishizu, H.; Fujita, A.; Tomoe, Y.; Siomi, M.C. Requirements for multivalent Yb body assembly in transposon silencing in Drosophila. EMBO Rep. 2019, 20, e47708.
- Qi, H.; Watanabe, T.; Ku, H.-Y.; Liu, N.; Zhong, M.; Lin, H. The Yb Body, a Major Site for Piwi-associated RNA Biogenesis and a Gateway for Piwi Expression and Transport to the Nucleus in Somatic Cells. J. Biol. Chem. 2011, 286, 3789–3797.
- Saito, K.; Ishizu, H.; Komai, M.; Kotani, H.; Kawamura, Y.; Nishida, K.M.; Siomi, H.; Siomi, M.C. Roles for the Yb body components Armitage and Yb in primary piRNA biogenesis in Drosophila. Genes Dev. 2010, 24, 2493–2498.
- Haase, A.D.; Fenoglio, S.; Muerdter, F.; Guzzardo, P.M.; Czech, B.; Pappin, D.J.; Chen, C.; Gordon, A.; Hannon, G.J. Probing the initiation and effector phases of the somatic piRNA pathway in Drosophila. Genes Dev. 2010, 24, 2499–2504.
- Olivieri, D.; Sykora, M.M.; Sachidanandam, R.; Mechtler, K.; Brennecke, J. An in vivo RNAi assay identifies major genetic and cellular requirements for primary piRNA biogenesis in Drosophila. EMBO J. 2010, 29, 3301–3317.
- Zamparini, A.L.; Davis, M.Y.; Malone, C.D.; Vieira, E.; Zavadil, J.; Sachidanandam, R.; Hannon, G.J.; Lehmann, R. Vreteno, a gonad-specific protein, is essential for germline development and primary piRNA biogenesis in Drosophila. Development 2011, 138, 4039–4050.
- Handler, D.; Olivieri, D.; Novatchkova, M.; Gruber, F.S.; Meixner, K.; Mechtler, K.; Stark, A.; Sachidanandam, R.; Brennecke, J. A systematic analysis of DrosophilaTUDOR domain-containing proteins identifies Vreteno and the Tdrd12 family as essential primary piRNA pathway factors. EMBO J. 2011, 30, 3977–3993.
- Pane, A.; Wehr, K.; Schüpbach, T. zucchini and squash Encode Two Putative Nucleases Required for rasiRNA Production in the Drosophila Germline. Dev. Cell 2007, 12, 851–862.
- Olivieri, D.; Senti, K.-A.; Subramanian, S.L.; Sachidanandam, R.; Brennecke, J. The Cochaperone Shutdown Defines a Group of Biogenesis Factors Essential for All piRNA Populations in Drosophila. Mol. Cell 2012, 47, 954–969.
- Preall, J.B.; Czech, B.; Guzzardo, P.M.; Muerdter, F.; Hannon, G.J. shutdown is a component of the Drosophila piRNA biogenesis machinery. RNA 2012, 18, 1446–1457.
- Horwich, M.D.; Li, C.; Matranga, C.; Vagin, V.; Farley, G.; Wang, P.; Zamore, P.D. The Drosophila RNA Methyltransferase, DmHen1, Modifies Germline piRNAs and Single-Stranded siRNAs in RISC. Curr. Biol. 2007, 17, 1265–1272.
- Malone, C.D.; Brennecke, J.; Dus, M.; Stark, A.; McCombie, W.R.; Sachidanandam, R.; Hannon, G.J. Specialized piRNA Pathways Act in Germline and Somatic Tissues of the Drosophila Ovary. Cell 2009, 137, 522–535.
- Gunawardane, L.S.; Saito, K.; Nishida, K.M.; Miyoshi, K.; Kawamura, Y.; Nagami, T.; Siomi, H.; Siomi, M.C. A Slicer-Mediated Mechanism for Repeat-Associated siRNA 5’ End Formation in Drosophila. Science 2007, 315, 1587–1590.
- Lim, A.K.; Kai, T. Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2007, 104, 6714–6719.
- Patil, V.S.; Kai, T. Repression of Retroelements in Drosophila Germline via piRNA Pathway by the Tudor Domain Protein Tejas. Curr. Biol. 2010, 20, 724–730.
- Cook, H.A.; Koppetsch, B.S.; Wu, J.; Theurkauf, W.E. The Drosophila SDE3 Homolog armitage Is Required for oskar mRNA Silencing and Embryonic Axis Specification. Cell 2004, 116, 817–829.
- Han, B.W.; Zamore, P.D. PiRNAs. Curr. Biol. 2014, 24, R730–R733.
- Calcagno, D.Q.; Mota, E.R.D.S.; Moreira, F.; De Sousa, S.B.M.; Burbano, R.R.; Assumpção, P.P.; Patel, V.; Preedy, V. Role of PIWI-Interacting RNA (piRNA) as Epigenetic Regulation. In Handbook of Nutrition, Diet, and Epigenetics; Springer: Berlin/Heidelberg, Germany, 2017; pp. 1–23.
- Gainetdinov, I.; Colpan, C.; Arif, A.; Cecchini, K.; Zamore, P.D. A Single Mechanism of Biogenesis, Initiated and Directed by PIWI Proteins, Explains piRNA Production in Most Animals. Mol. Cell 2018, 71, 775–790.
- Ayarpadikannan, S.; Kim, H.-S. The Impact of Transposable Elements in Genome Evolution and Genetic Instability and Their Implications in Various Diseases. Genom. Inform. 2014, 12, 98–104.
- Ng, K.W.; Anderson, C.S.; Marshall, E.A.; Minatel, B.C.; Enfield, K.S.S.; Saprunoff, H.L.; Lam, W.L.; Martinez, V.D. Piwi-interacting RNAs in cancer: Emerging functions and clinical utility. Mol. Cancer 2016, 15, 1–13.
- Chen, Y.; Pane, A.; Schüpbach, T. Cutoff and aubergine Mutations Result in Retrotransposon Upregulation and Checkpoint Activation in Drosophila. Curr. Biol. 2007, 17, 637–642.
- Sarot, E.; Payen-Groschêne, G.; Bucheton, A.; Pélisson, A. Evidence for a piwi-Dependent RNA Silencing of the gypsy Endogenous Retrovirus by the Drosophila melanogaster flamenco Gene. Genetics 2004, 166, 1313–1321.
- Rayford, K.J.; Cooley, A.; Arun, A.; Rachakonda, G.; Kleschenko, Y.; Villalta, F.; Pratap, S.; Lima, M.F.; Nde, P.N. Trypanosoma cruzi Modulates PIWI-Interacting RNA Expression in Primary Human Cardiac Myocytes during the Early Phase of Infection. Int. J. Mol. Sci. 2020, 21, 9439.
- Girard, A.; Hannon, G.J. Conserved themes in small-RNA-mediated transposon control. Trends Cell Biol. 2008, 18, 136–148.
- Biryukova, I.; Ye, T. Endogenous siRNAs and piRNAs derived from transposable elements and genes in the malaria vector mosquito Anopheles gambiae. BMC Genom. 2015, 16, 278.
- Vagin, V.V.; Klenov, M.S.; Kalmykova, A.; Stolyarenko, A.D.; Kotelnikov, R.N.; Gvozdev, V.A. The RNA interference proteins and vasa locus are involved in the silencing of retrotransposons in the female germline of Drosophila melanogaster. RNA Biol. 2004, 1, 53–57.
- Savitsky, M.; Kwon, D.; Georgiev, P.; Kalmykova, A.; Gvozdev, V. Telomere elongation is under the control of the RNAi-based mechanism in the Drosophila germline. Genes Dev. 2006, 20, 345–354.
- Pélisson, A.; Sarot, E.; Payen-Groscheêne, G.; Bucheton, A. A Novel Repeat-Associated Small Interfering RNA-Mediated Silencing Pathway Downregulates Complementary Sense Gypsy Transcripts in Somatic Cells of the Drosophila Ovary. J. Virol. 2006, 81, 1951–1960.
- Shpiz, S.; Kwon, D.; Uneva, A.; Kim, M.; Klenov, M.; Rozovsky, Y.; Georgiev, P.; Savitsky, M.; Kalmykova, A. Characterization of Drosophila Telomeric Retroelement TAHRE: Transcription, Transpositions, and RNAi-based Regulation of Expression. Mol. Biol. Evol. 2007, 24, 2535–2545.
- Li, C.; Vagin, V.V.; Lee, S.; Xu, J.; Ma, S.; Xi, H.; Seitz, H.; Horwich, M.D.; Syrzycka, M.; Honda, B.M.; et al. Collapse of Germline piRNAs in the Absence of Argonaute3 Reveals Somatic piRNAs in Flies. Cell 2009, 137, 509–521.
- Carmell, M.A.; Girard, A.; Van De Kant, H.J.G.; Bourc’His, D.; Bestor, T.H.; De Rooij, D.G.; Hannon, G.J. MIWI2 Is Essential for Spermatogenesis and Repression of Transposons in the Mouse Male Germline. Dev. Cell 2007, 12, 503–514.
- Huang, X.A.; Yin, H.; Sweeney, S.; Raha, D.; Snyder, M.; Lin, H. A Major Epigenetic Programming Mechanism Guided by piRNAs. Dev. Cell 2013, 24, 502–516.
- Yin, H.; Lin, H. An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nat. Cell Biol. 2007, 450, 304–308.
- Watanabe, T.; Cui, X.; Yuan, Z.; Qi, H.; Lin, H. MIWI2 targets RNAs transcribed from piRNA-dependent regions to drive DNA methylation in mouse prospermatogonia. EMBO J. 2018, 37, e95329.
- Pal-Bhadra, M.; Leibovtch, B.A.; Gandhi, S.G.; Rao, M.; Bhadra, U.; Birchler, J.A.; Elgin, S.C.R. Heterochromatic Silencing and HP1 Localization in Drosophila are Dependent on the RNAi Machinery. Science 2004, 303, 669–672.
- Pal-Bhadra, M.; Bhadra, U.; Birchler, J.A. RNAi Related Mechanisms Affect Both Transcriptional and Posttranscriptional Transgene Silencing in Drosophila. Mol. Cell 2002, 9, 315–327.
- Gomes, A.Q.; Nolasco, S.; Soares, H. Non-Coding RNAs: Multi-Tasking Molecules in the Cell. Int. J. Mol. Sci. 2013, 14, 16010–16039.
- Rajasethupathy, P.; Antonov, I.; Sheridan, R.; Frey, S.; Sander, C.; Tuschl, T.; Kandel, E.R. A Role for Neuronal piRNAs in the Epigenetic Control of Memory-Related Synaptic Plasticity. Cell 2012, 149, 693–707.
- Kuramochi-Miyagawa, S.; Watanabe, T.; Gotoh, K.; Totoki, Y.; Toyoda, A.; Ikawa, M.; Asada, N.; Kojima, K.; Yamaguchi, Y.; Ijiri, T.W.; et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 2008, 22, 908–917.
- Fu, A.; Jacobs, D.I.; Zhu, Y. Epigenome-wide analysis of piRNAs in gene-specific DNA methylation. RNA Biol. 2014, 11, 1301–1312.
- Fu, A.; Jacobs, D.I.; Hoffman, A.E.; Zheng, T.; Zhu, Y. PIWI-interacting RNA 021285 is involved in breast tumorigenesis possibly by remodeling the cancer epigenome. Carcinogenesis 2015, 36, 1094–1102.
- Zoch, A.; Auchynnikava, T.; Berrens, R.V.; Kabayama, Y.; Schopp, T.; Heep, M.; Vasiliauskaite, L.; Pere-Rico, Y.A.; Cook, A.G.; Shkumatava, A.; et al. SPOCD1 is an essential executor of piRNA-directed de novo DNA methylation. Nature 2020, 584, 635–639.
- Schöpp, T.; Zoch, A.; Berrens, R.V.; Auchynnikava, T.; Kabayama, Y.; Vasiliauskaitė, L.; Rappsilber, J.; Allshire, R.C.; O’Carroll, D. TEX15 is an essential executor of MIWI2-directed transposon DNA methylation and silencing. Nat. Commun. 2020, 11, 1–8.
- Mohn, F.; Handler, D.; Brennecke, J. piRNA-guided slicing specifies transcripts for Zucchini-dependent, phased piRNA biogenesis. Science 2015, 348, 812–817.
- Robine, N.; Lau, N.C.; Balla, S.; Jin, Z.; Okamura, K.; Kuramochi-Miyagawa, S.; Blower, M.D.; Lai, E.C. A Broadly Conserved Pathway Generates 3′UTR-Directed Primary piRNAs. Curr. Biol. 2009, 19, 2066–2076.
- Nishida, K.M.; Saito, K.; Mori, T.; Kawamura, Y.; Nagami-Okada, T.; Inagaki, S.; Siomi, H.; Siomi, M.C. Gene silencing mechanisms mediated by Aubergine piRNA complexes in Drosophila male gonad. RNA 2007, 13, 1911–1922.
- Vourekas, A.; Zheng, Q.; Alexiou, P.; Maragkakis, M.; Kirino, Y.; Gregory, B.D.; Mourelatos, Z. Mili and Miwi target RNA repertoire reveals piRNA biogenesis and function of Miwi in spermiogenesis. Nat. Struct. Mol. Biol. 2012, 19, 773–781.
- Lee, E.J.; Banerjee, S.; Zhou, H.; Jammalamadaka, A.; Arcila, M.; Manjunath, B.S.; Kosik, K.S. Identification of piRNAs in the central nervous system. RNA 2011, 17, 1090–1099.
- Peng, L.; Song, L.; Liu, C.; Lv, X.; Li, X.; Jie, J.; Zhao, D.; Li, D. piR-55490 inhibits the growth of lung carcinoma by suppressing mTOR signaling. Tumor Biol. 2015, 37, 2749–2756.
- Esposito, T.; Magliocca, S.; Formicola, D.; Gianfrancesco, F. piR_015520 Belongs to Piwi-Associated RNAs Regulates Expression of the Human Melatonin Receptor 1A Gene. PLoS ONE 2011, 6, e22727.
- Temme, C.; Simonelig, M.; Wahle, E. Deadenylation of mRNA by the CCR4—NOT complex in Drosophila: Molecular and developmental aspects. Front. Genet. 2014, 5, 143.
- Filipowicz, W.; Bhattacharyya, S.N.; Sonenberg, N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet. 2008, 9, 102–114.
- Grivna, S.T.; Pyhtila, B.; Lin, H. MIWI associates with translational machinery and PIWI-interacting RNAs (piRNAs) in regulating spermatogenesis. Proc. Natl. Acad. Sci. USA 2006, 103, 13415–13420.
- Ramat, A.; Garcia-Silva, M.-R.; Jahan, C.; Naït-Saïdi, R.; Dufourt, J.; Garret, C.; Chartier, A.; Cremaschi, J.; Patel, V.; Decourcelle, M.; et al. The PIWI protein Aubergine recruits eIF3 to activate translation in the germ plasm. Cell Res. 2020, 30, 421–435.
- Unhavaithaya, Y.; Hao, Y.; Beyret, E.; Yin, H.; Kuramochi-Miyagawa, S.; Nakano, T.; Lin, H. MILI, a PIWI-interacting RNA-binding Protein, Is Required for Germ Line Stem Cell Self-renewal and Appears to Positively Regulate Translation. J. Biol. Chem. 2009, 284, 6507–6519.
- Aravin, A.A.; Lagos-Quintana, M.; Yalcin, A.; Zavolan, M.; Marks, D.; Snyder, B.; Gaasterland, T.; Meyer, J.; Tuschl, T. The Small RNA Profile during Drosophila melanogaster Development. Dev. Cell 2003, 5, 337–350.
- Lau, N.C.; Seto, A.G.; Kim, J.; Kuramochi-Miyagawa, S.; Nakano, T.; Bartel, D.P.; Kingston, R.E. Characterization of the piRNA Complex from Rat Testes. Science 2006, 313, 363–367.
- Meister, G.; Tuschl, T. Mechanisms of gene silencing by double-stranded RNA. Nat. Cell Biol. 2004, 431, 343–349.
- Jinek, M.; Doudna, J.A. A three-dimensional view of the molecular machinery of RNA interference. Nat. Cell Biol. 2008, 457, 405–412.
- Kim, V.N.; Nam, J.-W. Genomics of microRNA. Trends Genet. 2006, 22, 165–173.
- Liu, B.; Li, J.; Cairns, M.J. Identifying miRNAs, targets and functions. Brief. Bioinform. 2014, 15, 1–19.
- Dana, H.; Chalbatani, G.M.; Mahmoodzadeh, H.; Karimloo, R.; Rezaiean, O.; Moradzadeh, A.; Mehmandoost, N.; Moazzen, F.; Mazraeh, A.; Marmari, V.; et al. Molecular Mechanisms and Biological Functions of siRNA. Int. J. Biomed. Sci. IJBS 2017, 13, 48–57.
- Lam, J.K.W.; Chow, M.Y.T.; Zhang, Y.; Leung, S.W.S. siRNA Versus miRNA as Therapeutics for Gene Silencing. Mol. Ther. Nucleic Acids 2015, 4, e252.
- Bartel, D.P. MicroRNAs: Target Recognition and Regulatory Functions. Cell 2009, 136, 215–233.
- Martinez, N.J.; Walhout, A.J.M. The interplay between transcription factors and microRNAs in genome-scale regulatory networks. Bioessays 2009, 31, 435–445.
- Place, R.F.; Li, L.-C.; Pookot, D.; Noonan, E.J.; Dahiya, R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc. Natl. Acad. Sci. USA 2008, 105, 1608–1613.
- Ni, W.J.; Leng, X.M. Dynamic miRNA-mRNA paradigms: New faces of miRNAs. Biochem. Biophys. Rep. 2015, 4, 337–341.
- Catalanotto, C.; Cogoni, C.; Zardo, G. MicroRNA in Control of Gene Expression: An Overview of Nuclear Functions. Int. J. Mol. Sci. 2016, 17, 1712.
- Ellison, M.; Mittal, M.; Chaudhuri, M.; Chaudhuri, G.; Misra, S. The role of the redox/miR-6855-3p/PRDX5A axis in reversing SLUG-mediated BRCA2 silencing in breast cancer cells. Cell Commun. Signal. 2020, 18, 1–16.
- Liu, H.; Lei, C.; He, Q.; Pan, Z.; Xiao, D.; Tao, Y. Nuclear functions of mammalian MicroRNAs in gene regulation, immunity and cancer. Mol. Cancer 2018, 17, 1–14.
- Aravin, A.A.; Hannon, G.J.; Brennecke, J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science 2007, 318, 761–764.
- Gou, L.-T.; Dai, P.; Yang, J.-H.; Xue, Y.; Hu, Y.-P.; Zhou, Y.; Kang, J.-Y.; Wang, X.; Li, H.; Hua, M.-M.; et al. Pachytene piRNAs instruct massive mRNA elimination during late spermiogenesis. Cell Res. 2014, 24, 680–700.
- Rouget, C.; Papin, C.; Boureux, A.; Meunier, A.-C.; Franco, B.; Robine, N.; Lai, E.C.; Pelisson, A.; Simonelig, M. Maternal mRNA deadenylation and decay by the piRNA pathway in the early Drosophila embryo. Nature 2010, 467, 1128–1132.
- Zhang, H.; Kolb, F.A.; Brondani, V.; Billy, E.; Filipowicz, W. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J. 2002, 21, 5875–5885.
- Rana, T.M. Illuminating the silence: Understanding the structure and function of small RNAs. Nat. Rev. Mol. Cell Biol. 2007, 8, 23–36.
- Orban, T.I.; Izaurralde, E. Decay of mRNAs targeted by RISC requires XRN1, the Ski complex, and the exosome. RNA 2005, 11, 459–469.
- La Greca, A.; Scarafía, M.A.; Cañás, M.C.H.; Pérez, N.; Castañeda, S.; Colli, C.; Möbbs, A.M.; Velazque, N.L.S.; Neiman, G.; Garate, X.; et al. PIWI-interacting RNAs are differentially expressed during cardiac differentiation of human pluripotent stem cells. PLoS ONE 2020, 15, e0232715.
- Law, P.T.-Y.; Qin, H.; Ching, A.K.-K.; Lai, K.P.; Na Co, N.; He, M.; Lung, R.W.-M.; Chan, A.W.-H.; Chan, T.-F.; Wong, N. Deep sequencing of small RNA transcriptome reveals novel non-coding RNAs in hepatocellular carcinoma. J. Hepatol. 2013, 58, 1165–1173.
- Rajan, K.S.; Ramasamy, S. Retrotransposons and piRNA: The missing link in central nervous system. Neurochem. Int. 2014, 77, 94–102.
- Vella, S.; Gallo, A.; Nigro, A.L.; Galvagno, D.; Raffa, G.M.; Pilato, M.; Conaldi, P.G. PIWI-interacting RNA (piRNA) signatures in human cardiac progenitor cells. Int. J. Biochem. Cell Biol. 2016, 76, 1–11.
- Di Giacomo, M.; Comazzetto, S.; Saini, H.; De Fazio, S.; Carrieri, C.; Morgan, M.; Vasiliauskaite, L.; Benes, V.; Enright, A.J.; O’Carroll, D. Multiple Epigenetic Mechanisms and the piRNA Pathway Enforce LINE1 Silencing during Adult Spermatogenesis. Mol. Cell 2013, 50, 601–608.
- Lucchinetti, E.; Feng, J.; Da Silva, R.; Tolstonog, G.V.; Schaub, M.C.; Schumann, G.G.; Zaugg, M. Inhibition of LINE-1 expression in the heart decreases ischemic damage by activation of Akt/PKB signaling. Physiol. Genom. 2006, 25, 314–324.
- Sugden, P.H.; Fuller, S.J.; Weiss, S.C.; Clerk, A. Glycogen synthase kinase 3 (GSK3) in the heart: A point of integration in hypertrophic signalling and a therapeutic target? A critical analysis. Br. J. Pharmacol. 2008, 153, S137–S153.
- Gao, X.-Q.; Zhang, Y.-H.; Liu, F.; Ponnusamy, M.; Zhao, X.-M.; Zhou, L.-Y.; Zhai, M.; Liu, C.-Y.; Li, X.-M.; Wang, M.; et al. The piRNA CHAPIR regulates cardiac hypertrophy by controlling METTL3-dependent N6-methyladenosine methylation of Parp10 mRNA. Nat. Cell Biol. 2020, 22, 1–13.
- Yang, J.; Xue, F.-T.; Li, Y.-Y.; Liu, W.; Zhang, S. Exosomal piRNA sequencing reveals differences between heart failure and healthy patients. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 7952–7961.
- Rajan, K.S.; Velmurugan, G.; Gopal, P.; Ramprasath, T.; Babu, D.V.; Krithika, S.; Jenifer, Y.C.; Freddy, A.; William, G.; Kalpana, K.; et al. Abundant and Altered Expression of PIWI-Interacting RNAs during Cardiac Hypertrophy. Heart Lung Circ. 2016, 25, 1013–1020.
- Chong, Z.Z.; Shang, Y.C.; Maiese, K. Cardiovascular Disease and mTOR Signaling. Trends Cardiovasc. Med. 2011, 21, 151–155.




