| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Rüdiger Rudolf | + 2277 word(s) | 2277 | 2020-03-31 13:07:04 | | | |
| 2 | Catherine Yang | -13 word(s) | 2264 | 2020-10-29 08:34:07 | | | | |
| 3 | Catherine Yang | -13 word(s) | 2264 | 2020-10-29 08:34:46 | | |
Video Upload Options
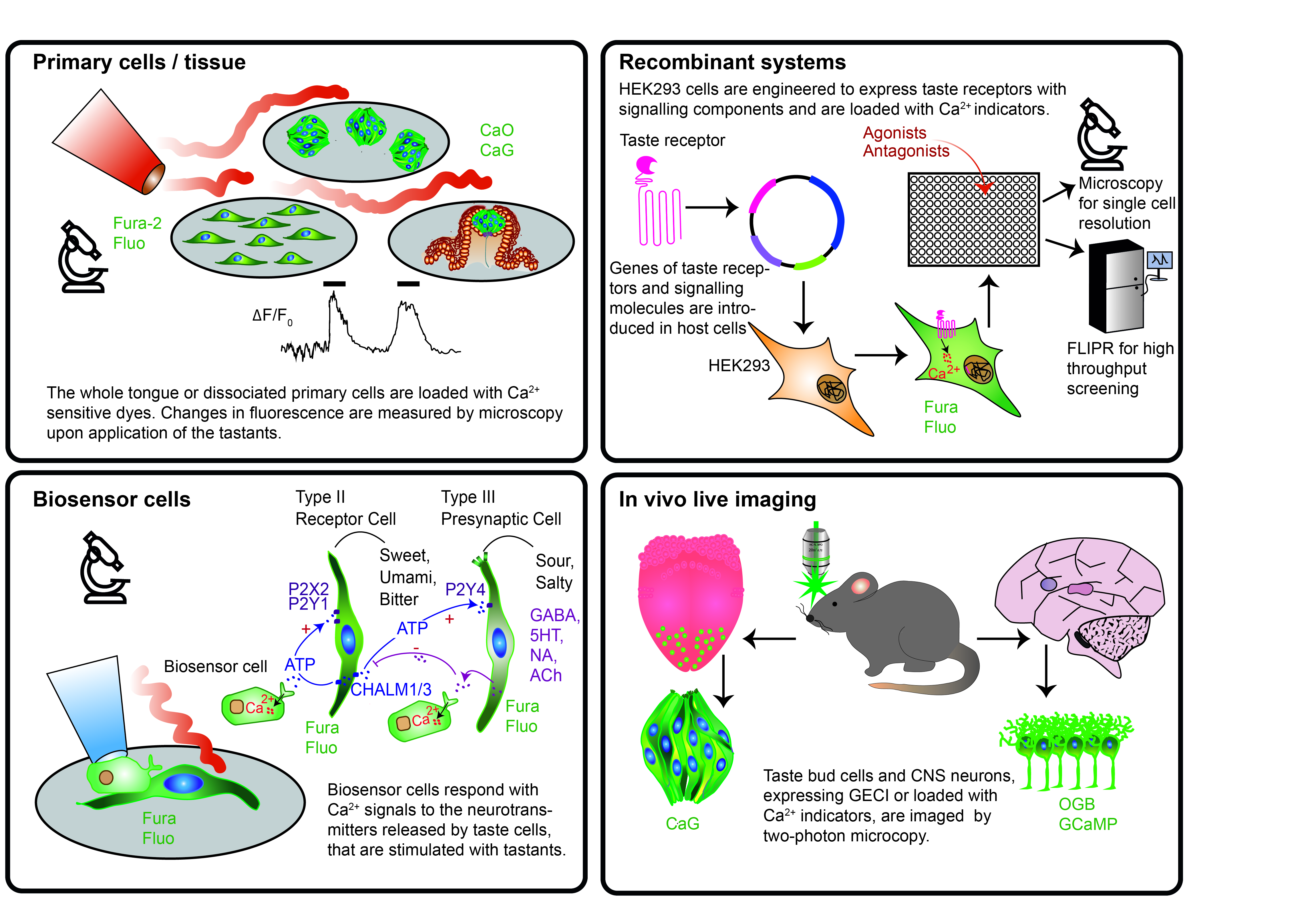
The five basic taste modalities, sweet, bitter, umami, salty and sour induce changes of Ca2+ levels, pH and/or membrane potential in taste cells of the tongue and/or in neurons that convey and decode gustatory signals to the brain. Optical biosensors, which can be either synthetic dyes or genetically encoded proteins whose fluorescence spectra depend on levels of Ca2+, pH or membrane potential, have been used in primary cells/tissues or in recombinant systems to study taste-related intra- and intercellular signaling mechanisms or to discover new ligands. Taste-evoked responses were measured by microscopy achieving high spatial and temporal resolution, while plate readers were employed for higher throughput screening. Here, these approaches making use of fluorescent optical biosensors to investigate specific taste-related questions or to screen new agonists/antagonists for the different taste modalities were reviewed systematically. Furthermore, in the context of recent developments in genetically encoded sensors, 3D cultures and imaging technologies, we propose new feasible approaches for studying taste physiology and for compound screening.
1. Introduction
On the journey towards understanding the sense of taste, two major parameters have been measured at the cellular level: voltage and Ca2+ changes. Indeed, both types of signals are emitted by taste cells in the periphery and by neurons in the central nervous system in response to a given tastant. All the way up from the tongue to the brain, our body uses an alphabet of voltage and Ca2+ fluctuations to decipher qualities and concentrations of tastants and to instruct decisions on food palatability. These notions have been used to construct and use different types of biosensors for fundamental and applied research.
Biosensors are analytical devices that allow the detection of chemical analytes through the combination of a biological recognition element with a physicochemical transducer [1][2]. Amongst these, cell-based biosensors incorporate sensors (and sometimes also transducers) in living cells and convert the resulting information into digital signals, which are then analyzed by means of specific techniques [2][3]. With respect to taste analysis, cell-based biosensors are also called “bioelectronic tongues” (B-ETs). These are able to analyze tastants in a fast, sensitive and selective way and have been recently designed based on the discoveries in taste physiology [2][4][5]. Since B-ETs are based on cells or tissues expressing taste receptors, they provide specificity, selectivity and sensitivity analogous to the biological sense of taste [5]. So far, transducers coupled to B-ETs use often voltametric/amperometric, potentiometric or piezoelectric methods and thus include microelectrode arrays, field effect transistors or light-addressable potentiometric sensors [5][6][7][8]. Even if these prototype devices have not yet been commercialized, B-ETs have a wide range of potential applications in the fields of drug and food industries, toxicology and biomedicine [3][9][10]. Furthermore, they might become valuable tools for research on taste transduction mechanisms. Finally, their development is tightly linked to the progress in understanding taste physiology.
A second kind of approach that has been primarily applied to basic research on taste sensation uses “molecular optical biosensors” to measure cellular responses to flavor stimulation. In this context, we refer to molecular optical biosensors as molecules that produce light changes in relation to cellular activity, therefore they are suitable to measure functional cell responses. They can be either, chemical fluorophores or genetically encoded fluorescent proteins, that shift their excitation/emission spectra or fluorescence intensity based on specific biological signals, such as changes in ion concentration, voltage, metabolites and second messenger molecules. Molecular optical biosensors are highly sensitive, real-time and simple-to-operate tools, that can be easily monitored with a plate reader for enhanced throughput or with microscopy for high spatial resolution. In contrast to classical B-ETs, molecular optical biosensors have been essential in basic research in the field of taste, but they have not yet been considered for the applications typical for B-ETs. This contribution reviews the use of molecular optical biosensors as tools in taste research and proposes their integration into the concept of B-ETs. Starting with a summary of the current knowledge on the taste system, the manuscript will systematically address applications of different molecular optical biosensors in the tongue, the central nervous system and in recombinant cell systems. Then, we will speculate on a future use of molecular optical biosensors to novel fields of applied taste research.
2. Conclusions and Future Perspectives
In taste research, two main streams run in parallel and are interconnected: one is the study of the mechanism underlying gustation, the other deals with the generation of artificial taste systems that can sense and discriminate between different tastants. Our concluding discussion will first focus on new potential approaches to discover the still missing pieces of the complex puzzle of taste physiology and to further refine the previous findings adding more details on the intracellular signaling and cell–cell communication. In a second part, ideas for novel sensing systems will be proposed that might mimic more closely the in vivo situation, but would be also compliant in terms of cost, technology and ethics.
3. New Approaches to Study Taste Physiology
The last 20 years have seen massive improvement in understanding taste physiology, both in the periphery and in the CNS as well as at the molecular, cellular and network levels. However, most studies focused on the ubiquitous intracellular signal Ca2+, potentially leading to an underestimation of other important factors. Indeed, the taste transduction pathways might be much more complex than the models proposed in most reviews including the pathway shown in Figure 1. For example, many knockout mouse models suggest that there may be additional signaling pathways involved, since taste sensation, nerve response and preferences for test compounds are often not completely abolished by eliminating a critical component of the canonical pathway including taste receptors, gustducin or TRPM5 channels [11][12][13][14]. Looking only at Ca2+ may not suffice to discriminate between these different pathways, as they may all induce a similar Ca2+ increase but with different underlying mechanisms. Along these lines, G-proteins other than gustducin are likely also involved in the signaling cascade [15][16] as well as other second messengers than Ca2+, such as cAMP [17][18]. Moreover, sweet signal transduction mechanisms may exist that bypass the activation of sweet taste receptors and instead use glucose transporters (GLUT) [12][19][20]. The pathway usage might be different even for compounds falling into the same taste modality [21], as it might be true for nutritive sugars and synthetic sweeteners or bitter compounds utilizing a PLCβ2-independent pathway at higher concentrations [22]. In this context, it would be helpful to use genetically encoded molecular biosensors for different signaling components such as cAMP and glucose [23][24] or a plethora of other factors (for a comprehensive database, see https://biosensordb.ucsd.edu). Via standard genetic engineering tools, such biosensors can be expressed in recombinant tongue-derived cell lines [25], introduced into primary taste cells via viral infection or electroporation or in transgenic mice for in situ or in vivo studies.
Next, it is widely recognized that specificity of second messenger-based cellular signal transduction involves local organization, by restricting signals to specific organelles and even to microdomains [26]. Until now, this aspect of signal compartmentation has been completely neglected in the investigation of taste transduction, although it is well known that taste cells use many different sources of Ca2+ [27][28]. Accordingly, although some taste responses are mediated by Ca2+ influx into receptor cells [29][30], the underlying molecular mechanism is unknown, since these cells do not express voltage-gated calcium channels. Finally, measurements of cytoplasmatic Ca2+ in combination with drugs that empty the endoplasmic reticulum or block the activity of mitochondria showed an important role of these organelles in the Ca2+ homeostasis within the taste bud [31][32]. As valuable as these insights were, future approaches should measure Ca2+ signals in the corresponding compartments by appropriate subcellular targeting of genetically encoded probes [33]. In synthesis, switching from chemical dyes to genetically encoded biosensors and adding other signaling components to Ca2+ will help to further improve our understanding of the taste signaling cascades.
4. Development of New In Vitro Taste Systems
Taste cell-based biosensors have a great potential for either basic research or for industrial applications, such as food safety and discovery of new ligands. On the one side, biosensor systems highly specialized for one taste modality are good testing platforms for discovering new agonists and antagonists to specific receptors/ions. Embedded into analytic devices they might serve to detect for example toxins [34] or to screen compound libraries [34][35]. On the other side, taste cell-based biosensors able to respond to flavors of different modalities could interpret complex flavor patterns and may be useful for quality control of food, drugs and the development of new palatable mixtures, such as in search for new sweet beverages with low caloric content.
On the long run, a complex taste cell-based artificial tongue system can be envisaged, which should fulfil the following criteria. It should 1) reproduce the complexity of the taste bud with respect to structure and functionality, 2) be representative for human gustation and 3) be implementable for high throughput. Considering the complexity of taste buds, 3D taste cell cultures with heterogeneous cell populations might come closest to such an idea. For reasons of reproducibility, these 3D culture could be formed as spheroids [25], cell chip cultures (Figure 2), and organoids derived from progenitor cells [36] or microfluidic devices [37]. Spheroids, prepared with tongue-derived human cells (HTC-8) by liquid overlay technique on commercially available low-attachment plates, were successfully used to measure real-time responses upon application of bitter compounds [25]. Comparable results were obtained upon 3D cultivation of HTC-8-G-GECO cells in special Dynarray cell chips, which can be used for live imaging experiments in perfusion conditions, as they are made of transparent materials and have porous walls (Figure 2, original data from us). Furthermore, the chips can be used in bioreactors with circulating media and with the option of different apical and basal perfusions to favor cell polarization. Further, organoids were generated from Lgr5-positive mouse progenitor cells, which could be expanded and differentiated into the different taste cell types [36][38]. However, neither the purification of such multipotent cells from human tongue, nor the generation of mature taste cells by differentiating human induced pluripotent stem cells have been explored so far. Recent progress in the mechanisms of taste cell development [36][39][40] suggest further steps in that direction. Another effort to recreate a multicellular taste cell-based system with a potential integration in a microfluid device for industrial application, has been recently made with the help of tongue extracellular matrix [37], which was generated from porcine tongue and used to prepare primary 2D and 3D taste cell cultures and co-cultures with neurons [37]. This improved both the expression of gustatory markers and the functionality of taste cells, furthermore taste cell signals were transmitted to the neurons. These findings were applied to test wine [10].
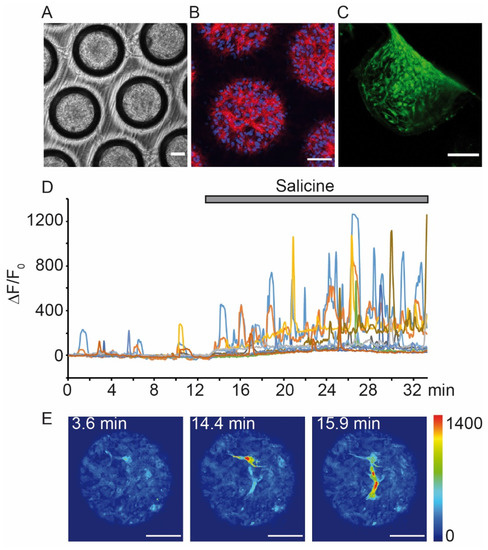
Figure 2. Use of a 3D culture with immortalized human tongue cells expressing a GECI to study Ca2+ responses upon perfusion of a bitter compound. Dynarray chips (300 Microns) were filled with 5000 HTC-8-G-GECO cells/cavity and cultured for three (A–C) or seven days (D,E). (A) Bright-field image. (B) Representative confocal fluorescence image of one optical plane at 30 µm from the top of the chip. Upon fixation, cells were labelled with markers for f-actin (phalloidin, red) and nuclei (DAPI, blue). (C) Confocal micrograph showing a side view of a chip cavity. Before microscopy, the chip was PFA-fixed and sliced. (D) Representative time course of HTC-8-G-GECO fluorescence changes (ΔF/F0, normalized to the time before compound perfusion). The chip was mounted in an Ibidi perfusion slide, stabilized with gelatine pads (Scaffolene, from Freudenberg) and then perfused with salicine (20 mM) (all procedures analogous to [25]). Traces show G-GECO fluorescence changes as a function of time, the time period of salicine perfusion is indicated by a grey bar. (E) Examples of pseudo-colored confocal images during perfusion in control (left panel) and in salicine solutions (middle and right panels). Blue and yellow/red cues indicate low and high G-GECO fluorescence intensities, respectively. Time points of image acquisition correspond to graph in (D). Images were acquired with an inverted Leica TCS SP8 confocal microscope. Scale bars in (A–C) and (E), 100 µm. (Unpublished data E.M. and T.C.)
Finally, on the road to an artificial tongue system, the development of optimal cell culture conditions needs to be flanked by novel sensor technologies for more specific and comprehensive readouts. For example, optical and electrical sensors could be used in parallel. Patterns of Ca2+ changes in combination with voltage current recordings may allow to discriminate not only between the different taste modalities but also between compounds of the same families, such as natural versus artificial sweeteners. Novel, sophisticated imaging systems suitable to acquire multiple fluorescence signals from microplates with cellular resolution are now available and high-content imaging systems allow the acquisition of confocal images in thick tissue/probes up to a 1536 wells format with multiple lasers and dichroic filters [41][42]. In addition, optical tissue clearing protocols start also to be applied to 3D cultures, which could largely improve expression analysis in three dimensions [43]. For enhanced throughput of electrical recordings, new devices such as multielectrode arrays or automatic patch-clamp platforms allow to clamp hundreds of cells simultaneously [44].
In summary, the use of taste cells to develop biosensor systems for gustation is a new field with a wide range of possible applications [5][6][7][9][45]. Next-generation biosensor systems could now be developed with 3D taste cell cultures that can be used for both high-content imaging and live imaging microscopy. The use of genetically encoded biosensors to measure different signaling molecules in defined cellular compartments will allow higher flexibility, easier handling and more specific readouts. A major challenge will be to couple 3D cell cultures expressing such sensors to microdevices able to read optical and electrical signals, in order to develop portable and user friendly “pocket tongues”.
References
- Ali, J.; Najeeb, J.; Asim Ali, M.; Farhan Aslam, M.; Raza, A. Biosensors: Their Fundamentals, Designs, Types and Most Recent Impactful Applications: A Review. J. Biosens. Bioelectron. 2017, 8.
- Liu, Q.; Wu, C.; Cai, H.; Hu, N.; Zhou, J.; Wang, P. Cell-based biosensors and their application in biomedicine. Chem. Rev. 2014, 114, 6423–6461.
- Podrażka, M.; Bączyńska, E.; Kundys, M.; Jeleń, P.S.; Witkowska Nery, E. Electronic Tongue-A Tool for All Tastes? Biosensors (Basel) 2017, 8, 3.
- Ha, D.; Sun, Q.; Su, K.; Wan, H.; Li, H.; Xu, N.; Sun, F.; Zhuang, L.; Hu, N.; Wang, P. Recent achievements in electronic tongue and bioelectronic tongue as taste sensors. Sens. Actuators B Chem. 2015, 207, 1136–1146.
- Wu, C.; Lillehoj, P.B.; Wang, P. Bioanalytical and chemical sensors using living taste, olfactory, and neural cells and tissues: A short review. Analyst 2015, 140, 7048–7061.
- Wu, C.; Du, Y.-W.; Huang, L.; Ben-Shoshan Galeczki, Y.; Dagan-Wiener, A.; Naim, M.; Niv, M.Y.; Wang, P. Biomimetic Sensors for the Senses: Towards Better Understanding of Taste and Odor Sensation. Sensors (Basel) 2017, 17, 2881.
- Wasilewski, T.; Kamysz, W.; Gębicki, J. Bioelectronic tongue: Current status and perspectives. Biosens. Bioelectron. 2020, 150, 111923.
- Bioinspired Smell and Taste Sensors, 1st ed.; Wang, P.; Liu, Q.; Wu, C.; Hsia, K.J. (Eds.) Springer: Dordrecht, The Netherlands; Beijing, China, 2015; ISBN 9789401773324.
- Woertz, K.; Tissen, C.; Kleinebudde, P.; Breitkreutz, J. Taste sensing systems (electronic tongues) for pharmaceutical applications. Int. J. Pharm. 2011, 417, 256–271.
- Gutiérrez, M.; Llobera, A.; Vila-Planas, J.; Capdevila, F.; Demming, S.; Büttgenbach, S.; Mínguez, S.; Jiménez-Jorquera, C. Hybrid electronic tongue based on optical and electrochemical microsensors for quality control of wine. Analyst 2010, 135, 1718–1725.
- Wong, G.T.; Gannon, K.S.; Margolskee, R.F. Transduction of bitter and sweet taste by gustducin. Nature 1996, 381, 796–800.
- Damak, S.; Rong, M.; Yasumatsu, K.; Kokrashvili, Z.; Varadarajan, V.; Zou, S.; Jiang, P.; Ninomiya, Y.; Margolskee, R.F. Detection of sweet and umami taste in the absence of taste receptor T1r3. Science 2003, 301, 850–853.
- Damak, S.; Mosinger, B.; Margolskee, R.F. Transsynaptic transport of wheat germ agglutinin expressed in a subset of type II taste cells of transgenic mice. BMC Neurosci. 2008, 9, 96.
- Glendinning, J.I.; Bloom, L.D.; Onishi, M.; Zheng, K.H.; Damak, S.; Margolskee, R.F.; Spector, A.C. Contribution of α-Gustducin to Taste-guided Licking Responses of Mice. Chem Senses 2005, 30, 299–316.
- Tizzano, M.; Dvoryanchikov, G.; Barrows, J.K.; Kim, S.; Chaudhari, N.; Finger, T.E. Expression of Galpha14 in sweet-transducing taste cells of the posterior tongue. BMC Neurosci. 2008, 9, 110.
- Ruiz-Avila, L.; McLaughlin, S.K.; Wildman, D.; McKinnon, P.J.; Robichon, A.; Spickofsky, N.; Margolskee, R.F. Coupling of bitter receptor to phosphodiesterase through transducin in taste receptor cells. Nature 1995, 376, 80–85.
- Clapp, T.R.; Trubey, K.R.; Vandenbeuch, A.; Stone, L.M.; Margolskee, R.F.; Chaudhari, N.; Kinnamon, S.C. Tonic activity of Gα-gustducin regulates taste cell responsivity. FEBS Lett. 2008, 582, 3783–3787.
- Trubey, K.R.; Culpepper, S.; Maruyama, Y.; Kinnamon, S.C.; Chaudhari, N. Tastants evoke cAMP signal in taste buds that is independent of calcium signaling. Am. J. Physiol. Cell Physiol. 2006, 291, C237–C244.
- Yee, K.K.; Sukumaran, S.K.; Kotha, R.; Gilbertson, T.A.; Margolskee, R.F. Glucose transporters and ATP-gated K+ (KATP) metabolic sensors are present in type 1 taste receptor 3 (T1r3)-expressing taste cells. Proc. Natl. Acad. Sci. USA 2011, 108, 5431–5436.
- Glendinning, J.I.; Frim, Y.G.; Hochman, A.; Lubitz, G.S.; Basile, A.J.; Sclafani, A. Glucose elicits cephalic-phase insulin release in mice by activating KATP channels in taste cells. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2017, 312, R597–R610.
- Bernhardt, S.J.; Naim, M.; Zehavi, U.; Lindemann, B. Changes in IP3 and cytosolic Ca2+ in response to sugars and non-sugar sweeteners in transduction of sweet taste in the rat. J. Physiol. (Lond) 1996, 490, 325–336.
- Dotson, C.D.; Roper, S.D.; Spector, A.C. PLCbeta2-independent behavioral avoidance of prototypical bitter-tasting ligands. Chem Senses 2005, 30, 593–600.
- Zaccolo, M.; Giorgi, F.D.; Cho, C.Y.; Feng, L.; Knapp, T.; Negulescu, P.A.; Taylor, S.S.; Tsien, R.Y.; Pozzan, T. A genetically encoded, fluorescent indicator for cyclic AMP in living cells. Nat. Cell Biol. 2000, 2, 25–29.
- Deuschle, K.; Okumoto, S.; Fehr, M.; Looger, L.L.; Kozhukh, L.; Frommer, W.B. Construction and optimization of a family of genetically encoded metabolite sensors by semirational protein engineering. Protein Sci. 2005, 14, 2304–2314.
- Von Molitor, E.; Nürnberg, E.; Ertongur-Fauth, T.; Scholz, P.; Riedel, K.; Hafner, M.; Rudolf, R.; Cesetti, T. Analysis of Calcium Signaling in Live Human Tongue Cell 3D-Cultures upon Tastant Perfusion. Cell Calcium 2020, 102164.
- Filadi, R.; Pozzan, T. Generation and functions of second messengers microdomains. Cell Calcium 2015, 58, 405–414.
- DeFazio, R.A.; Dvoryanchikov, G.; Maruyama, Y.; Kim, J.W.; Pereira, E.; Roper, S.D.; Chaudhari, N. Separate populations of receptor cells and presynaptic cells in mouse taste buds. J. Neurosci. 2006, 26, 3971–3980.
- Clapp, T.R.; Medler, K.F.; Damak, S.; Margolskee, R.F.; Kinnamon, S.C. Mouse taste cells with G protein-coupled taste receptors lack voltage-gated calcium channels and SNAP-25. BMC Biol. 2006, 4, 7.
- Hacker, K.; Laskowski, A.; Feng, L.; Restrepo, D.; Medler, K. Evidence for two populations of bitter responsive taste cells in mice. J. Neurophysiol. 2008, 99, 1503–1514.
- Hochheimer, A.; Krohn, M.; Rudert, K.; Riedel, K.; Becker, S.; Thirion, C.; Zinke, H. Endogenous gustatory responses and gene expression profile of stably proliferating human taste cells isolated from fungiform papillae. Chem. Senses 2014, 39, 359–377.
- Rebello, M.R.; Maliphol, A.B.; Medler, K.F. Ryanodine Receptors Selectively Interact with L Type Calcium Channels in Mouse Taste Cells. PLoS ONE 2013, 8, e68174.
- Rebello, M.R.; Maliphol A., B.; Medler, K.F. Ryanodine Receptors Selectively Interact with L Type Calcium Channels in Mouse Taste Cells. PLoS ONE 2013, 8, e68174.
- Mehta, S.; Zhang, J. Dynamic visualization of calcium-dependent signaling in cellular microdomains. Cell Calcium 2015, 58, 333–341.
- Wu, C.; Du, L.; Zou, L.; Zhao, L.; Huang, L.; Wang, P. Recent advances in taste cell- and receptor-based biosensors. Sens. Actuators B Chem. 2014, 201, 75–85.
- Slack, J.P.; Brockhoff, A.; Batram, C.; Menzel, S.; Sonnabend, C.; Born, S.; Galindo, M.M.; Kohl, S.; Thalmann, S.; Ostopovici-Halip, L.; et al. Modulation of bitter taste perception by a small molecule hTAS2R antagonist. Curr. Biol. 2010, 20, 1104–1109.
- Ren, W.; Lewandowski, B.C.; Watson, J.; Aihara, E.; Iwatsuki, K.; Bachmanov, A.A.; Margolskee, R.F.; Jiang, P. Single Lgr5- or Lgr6-expressing taste stem/progenitor cells generate taste bud cells ex vivo. Proc. Natl. Acad. Sci. USA 2014, 111, 16401–16406.
- Lee, J.S.; Cho, A.-N.; Jin, Y.; Kim, J.; Kim, S.; Cho, S.-W. Bio-artificial tongue with tongue extracellular matrix and primary taste cells. Biomaterials 2018, 151, 24–37.
- Aihara, E.; Mahe, M.M.; Schumacher, M.A.; Matthis, A.L.; Feng, R.; Ren, W.; Noah, T.K.; Matsu-ura, T.; Moore, S.R.; Hong, C.I.; et al. Characterization of stem/progenitor cell cycle using murine circumvallate papilla taste bud organoid. Sci. Rep. 2015, 5, 17185.
- Barlow, L.A. Progress and renewal in gustation: New insights into taste bud development. Development 2015, 142, 3620–3629.
- Ren, W.; Aihara, E.; Lei, W.; Gheewala, N.; Uchiyama, H.; Margolskee, R.F.; Iwatsuki, K.; Jiang, P. Transcriptome analyses of taste organoids reveal multiple pathways involved in taste cell generation. Sci. Rep. 2017, 7, 4004.
- Chessel, A.; Carazo Salas, R.E. From observing to predicting single-cell structure and function with high-throughput/high-content microscopy. Essays Biochem. 2019, 63, 197–208.
- Booij, T.H.; Price, L.S.; Danen, E.H.J. 3D Cell-Based Assays for Drug Screens: Challenges in Imaging, Image Analysis, and High-Content Analysis. SLAS Discov. 2019, 24, 615–627.
- Nürnberg, E.; Vitacolonna, M.; Klicks, J.; Von Molitor, E.; Cesetti, T.; Keller, F.; Bruch, R.; Ertongur-Fauth, T.; Riedel, K.; Scholzt, P.; et al. Routine Optical Clearing of 3D-Cell Cultures: Simplicity Forward. Front. Mol. Biosci. 2020, 7, 20.
- Li, T.; Lu, G.; Chiang, E.Y.; Chernov-Rogan, T.; Grogan, J.L.; Chen, J. High-throughput electrophysiological assays for voltage gated ion channels using SyncroPatch 768PE. PLoS ONE 2017, 12, e0180154.
- Wu, A.; Dvoryanchikov, G.; Pereira, E.; Chaudhari, N.; Roper, S.D. Breadth of tuning in taste afferent neurons varies with stimulus strength. Nat. Commun. 2015, 6, 8171.