The bromodomain adjacent to the zinc finger domain 1B (BAZ1B) or Williams syndrome transcription factor (WSTF) are just two of the names referring the same protein that is encoded by the WBSCR9 gene and is among the 26–28 genes that are lost from one copy of 7q11.23 in Williams syndrome (WS: OMIM 194050). Patients afflicted by this contiguous gene deletion disorder present with a range of symptoms including cardiovascular complications, developmental defects as well as a characteristic cognitive and behavioral profile. Studies in patients with atypical deletions and mouse models support BAZ1B hemizygosity as a contributing factor to some of the phenotypes. Focused analysis on BAZ1B has revealed this to be a versatile nuclear protein with a central role in chromatin remodeling through two distinct complexes as well as being involved in the replication and repair of DNA, transcriptional processes involving RNA Polymerases I, II, and III as well as possessing kinase activity.
1. Introduction
The Williams syndrome transcription factor (WSTF) encoded by the WBSCR9 gene was first reported in 1998 by two independent studies
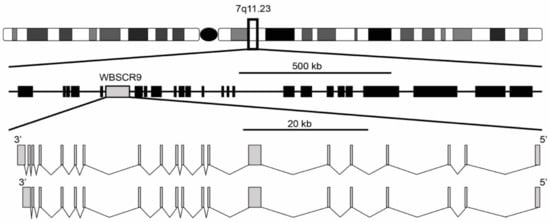
[1][2]. It was described as a single copy gene spanning 80 kb with a 4449 bp open reading frame encoding a protein of 1483 amino acids. The gene structure is composed of two isoforms with either 19 or 20 exons, with the 20-exon isoform differing by splicing to an extra 3′ non-coding exon that increases the size of the 3′ untranslated region (
Figure 1). Exons range from 99 bp for exon 11 to 1702 bp for exon 7 and are flanked by the standard GT-AG splice donor and acceptor sequences. The gene is located at 7q11.23 and is oriented 5′ telomeric and 3′ centromeric. Through the use of florescence in situ hybridization, WBSCR9 was physically mapped to the Williams syndrome (WS) deletion region
[1]. Williams Syndrome is a contiguous gene deletion syndrome (OMIM 194050) that causes a complex developmental disorder that presents with multi-systemic defects. Intellectual disability, dysmorphic facial features, a unique cognitive profile along with congenital cardiovascular complications, infantile hypercalcemia, and growth deficiency are common characteristics of WS patients
[3][4]. The syndrome results from a heterozygous deletion of >1 Mb (up to 1.83 Mb) mapping to 7q11.23 (
Figure 1), which is prone to non-allelic homologous recombination and encompasses roughly 28 genes
[5][6]. Duplication of the same interval in 7q11.23 Duplication Syndrome (OMIM 609757) results in individuals being triploid for most of the same genes and is characterized by numerous phenotypes including cardiac abnormalities, craniofacial developmental defects, anxiety, and autistic spectrum disorder
[7].
Figure 1. Genomic organization of WS deletion region including WBSCR9 gene. There are two isoforms of BAZ1B gene on DNA minus strand with either 19 or 20 exons.
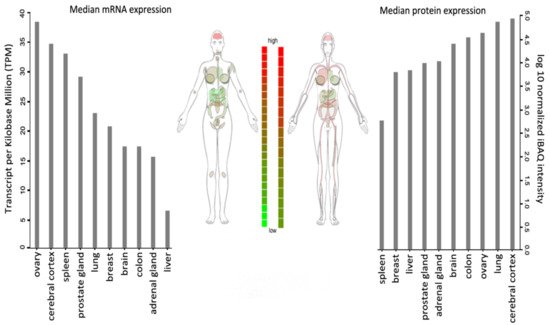
WSTF is universally expressed, with the highest transcript levels being detected in adult brain, heart, ovary, placenta, and skeletal muscle tissues (Figure 2).
Figure 2. BAZ1B expression in different tissues. Images and expression data were obtained from proteomics database
[8][9]. Body visualization is a female body. Proteomics data comes from mass spectrometry (MS1) analysis. Transcriptomics data comes from RNASeq analysis.
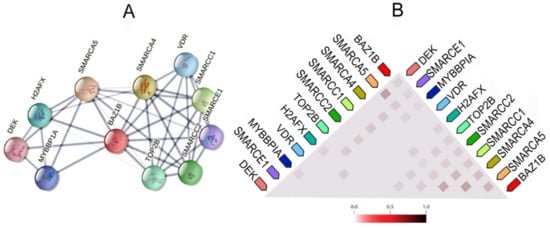
It also forms physical associations with several other important proteins. Figure 3 illustrates the network of the functional and physical associations between BAZ1B and the first shell of interactors as well as their observed co-expression in Homo sapiens (Figure 3).
Figure 3. BAZ1B co-expression and interactions. Figures are obtained from STRING database
[10]. (
A) Each node in the network represents all proteins produced by a single, protein-coding gene locus, i.e., splice isoforms or posttranslational modifications are collapsed. Edges represent protein–protein association, which are meant to be specific and meaningful. Proteins jointly contribute to a shared function; however, this does not necessarily mean they are physically binding each other. Line thickness indicates the strength of data support. (
B) Co-expression of BAZ1B and first shell of interactors. The color intensity in the triangular matrices indicates the level of confidence that two proteins are functionally associated, given the overall expression data in the organism.
The WSTF protein has a molecular mass of 171 kDa and was speculated by Lu et al. to function as a transcription factor based upon the existence of conserved motifs in the protein sequence (
Figure 4). In addition to the conserved motifs, several phosphorylation, amidation, 13 N-myristylation, and glycosylation sites as well as a specific acidic amino acid-enriched region (amino acids 1260–1274) were annotated
[1]. Additionally, there is a polyglutamate stretch (13 residues) and several nuclear localization signals (three types) that support the protein to function in the nucleus. Northern blot hybridization analysis has also revealed an alternatively spliced smaller 4.5 kb message that excludes the 1.7-kb exon 7 with comparable expression levels to the larger transcript in the liver, lung, and kidney. However, the larger transcript is predominantly expressed in the brain and placenta
[2].
2. Bromodomain
Within the C-terminus of WSTF is a bromodomain (BrD) that spans 71 amino acids from residues 1356 to 1426, contains a consensus sequence and helix-turn-helix structure, and is required for protein–protein interactions
[1][2] (
Figure 4).
The BrD is an evolutionary conserved structural domain in proteins that bind acetylated lysine (Kac) residues
[11][12][13]. This interaction is mediated by a highly conserved Asparagine residue within the BrD. For instance, it has been demonstrated through the crystal structure that the formation of a hydrogen bond between the amide nitrogen of Asn407 (the conserved asparagine in the BrD of Saccharomyces cerevisiae Gen5p bromodomain complex) and the oxygen of the acetyl carbonyl group is a key event in the BrD-mediated protein–protein interaction between Gen5p and histone H4 acetylated at lysine 16
[14]. BrD proteins mediate protein–protein interactions in DNA recombination, replication, and repair; in histone modifications and chromatin remodeling; as well as in the recruitment of transcription factors that affect both transcription initiation and elongation processes
[15]. The inhibition of BrD and extra-terminal domain (BET) family proteins prevents proliferation in carcinoma cells
[16] and blocks the expression of inflammatory genes
[17].
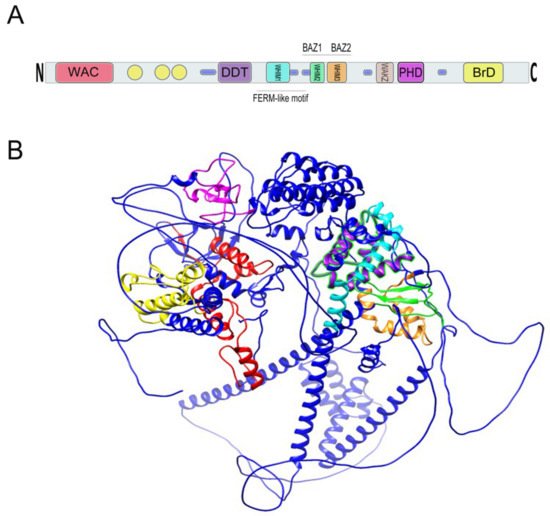
Figure 4. (
A) Relative location of BAZ1B domains. Yellow circles are low complexity regions. Blue cylinders are coiled coil regions. The first N-terminally located domain is present in Acf1-related proteins in a variety of organisms. The WAC (WSTF/Acf1/cbp146) domain is within the DNA binding region of the Acf1 and is believed to be involved in DNA binding and is associated with the PHD finger and Brd domains in other proteins
[18][19]. It spans residues 21–120 on BAZ1B. DDT stands for DNA-binding homobox-containing proteins and Different Transcription and chromatin remodeling factors. Such proteins share this domain, which spans almost 60 residues and is associated exclusively with nuclear domains including PHD finger, Brd, and DNA-binding homeodomain
[20]. It spans residues 604–668 on BAZ1B. Residues 724–773 on BAZ1B locate WHIM1 conserved the
α helical motif that along with the WHIM2, WHIM3, and DDT domain comprise an
α helical module that is reported to interact with linker DNA and the SLIDE domain of ISWI proteins
[21]. WHIM2 domain spans residues 899 to 936 on BAZ1B and contains the D-TOX E motif that is also known as the Williams–Beuren Syndrome DDT (WSD) motif. It is conserved from yeast to animals
[22]. PHD zinc finger (spanning 1184 to 1234) and bromodomain (1356–1426) are the two C-terminally located domains. Refer to the main text for more information about these domains. (
B) A 3D representation of BAZ1B based on its PDB file (PDB ID: Q9UIG0) predicted by the AlphaFold protein structure database
[23]. Domains are highlighted with different colors as follows: BrD: yellow; PHD: magenta; WHIM1: cyan; WHIM2: green; WHIM3: orange; DDT: purple; WAC: red.
3. Plant Homeodomain Finger
The second conserved motif directly adjacent to the BrD is a cysteine/histidine-rich plant homeodomain type (PHD-type) zinc finger motif (
Figure 4) that encompasses 51 residues (1184–1234) and that contains the conserved characteristics of a typical PHD finger
[1][2].
PHD fingers are structurally conserved zinc-finger-like motifs with a unique Cys4-His-Cys3 pattern that is distinct from that of similar sized RING finger and LIM domain zinc fingers
[24] (
Figure 5). These versatile epigenome readers interact with the first six N-terminal residues of histone H3. Several studies have implicated intriguing and complicated functions of PHD fingers in reading histone codes such as histone H3 tri-methylated at lysine 4 (H3K4me3), unmethylated (H3K4me0), arginine 2 of histone H3 (H3R2), and histone H3 acetylated at lysine 14 (H3K14ac). A single PHD finger can even recognize a combination of histone post-translation modifications by using multiple binding sites within the motif. For example, double tandem PHD Finger protein 3b (DPF3b), reads the combination of H3K14ac, H3K4me0 and H3R2
[25]. In addition to the BrD, PHD fingers are highlighted as epigenetic modulators of transcription via the recruitment of chromatin remodeling and transcription factors as well as basal transcription machinery
[12][24][25].
Figure 5. Images are obtained from the protein data bank. The structure information comes from primary publication
[26]. (
A) Tertiary structure of 51 residues in PHD zinc finger domain of BAZ1B with the two zinc ions (Zn
2+) (blue sticks) are shown (residues 1185–1235). PDB ID: 1F62. (
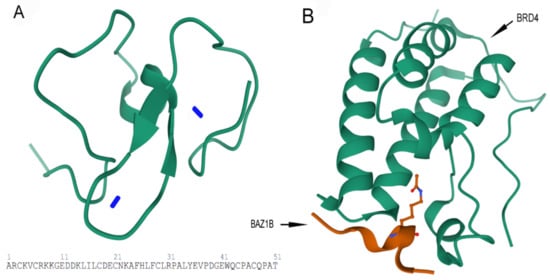
B) Crystal structure of first bromodomain of human BRD4 in a complex with an acetylated BAZ1B peptide (residues 217–226; FLPH(ALY)YDVKL; K221 ac). PDB ID: 5NNF.
4. WAKZ and WAC Motifs
In addition to the BrD and PHD finger, there is a WAKZ (WSTF/Acf1/KIAA0314/ZK783.4) motif located immediately proximal to the PHD finger as well as a WAC (WSTF/Acf1/cbp146) motif at the amino terminus of WSTF that spans 107 residues (20–126) (
Figure 4). These two motifs were first described by Ito et al. in the ATP-dependent chromatin assembly factor 1 (Acf1) protein of
Drosophila melanogaster. They described Acf1 as a novel protein containing two PHD fingers, a bromodomain, a WAKZ, and a WAC domain
[18].
WAKZ is a common motif between members of the highly conserved bromodomain adjacent zinc finger, the BAZ family in humans, as well as their homolog in
Drosophila (Acf1) and
Caenorhabditis (ZK783.4)
[13]. Chromatin assembly is slightly defective when the WAKZ, PHD finger, and BrD domains are mutated in Acf1; however, the binding of the ACF complex (Acf1/ISWI) to DNA is not impaired upon the mutation of either of these regions
[19]. In addition to the WAKZ motif, the WAC motif located in the N-terminal is also a shared feature between members of the BAZ family and Acf1
[13]. Concordantly, Poot et al. identified these proteins as the WAL (WSTF/Acf1 Like) family. WAC motifs are required for DNA binding and may be sufficient for WSTF to target heterochromatin in pericentric regions
[19]. The WAC domain in conjunction with the adjacent C-Motif (amino acids 206–345) have also been reported to function as a tyrosine kinase
[27] (See
Section 3.1 below).
5. WSTF or BAZ1B?
The BAZ gene family was first described as a novel BrD family by Jones et al. that has two subfamilies when considering their conserved residues. The BAZ1 subfamily involves BAZ1A (14q12-q13) and BAZ1B (7q11-q21), whereas the BAZ2 subfamily is comprised of BAZ2A (12q24.2-qter) and BAZ2B (2q23-q24). Based on a high degree of sequence conservation, the BAZ1 subfamily is closely related to Acf1, and the BAZ2 subfamily is related to the baz-2 (ZK783.4) of
Caenorhabditis elegans and are considered orthologues
[13]. BAZ1B is more related to human Acf1 (hACF1 or BAZ1A) than BAZ2A and BAZ2B are. The common WAC motif at the N-terminus of the proteins is also another distinct feature of the BAZ1 subfamily compared to more distant members of the WAL family
[28]. Northern analysis has confirmed the expression of approximately 7.5 kb and 9.5 kb transcripts of BAZ1 and BAZ2 genes, respectively, in a range of tissues with markedly variable expression levels. For instance, BAZ1A is highly expressed in the testis and BAZ2A is moderately expressed in most analyzed tissues. This might confer tissue-specific transcriptional regulatory functions. All BAZ members have seven highly conserved motifs, out of which six are shared among all BAZ members. The WAC domain is present in BAZ1/Acf1 proteins but not in BAZ2/ZK783.4; on the other hand, a ZB2 domain is shared between BAZ2/ZK783.4 but not BAZ1/Acf1 proteins. BAZ1B, the second member of the BAZ1 subfamily reported by Jones et al., is WSTF and, therefore, from this point forward, WSTF shall be referred to as BAZ1B for the remainder of this review.
6. BAZ or WHIM Motifs?
In addition to the abovementioned BrD, PHD finger, WAKZ, and WAC motifs, Jones et al. also identified BAZ1 and BAZ2 (
Figure 4) as two other conserved motifs between all BAZ members as well as the Acf1 and Zk783.4 proteins
[13]. More than a decade later, Aravind and Iyer also reported a novel N-terminal domain in the human ASLX protein and named it HARE-helix-turn-helix (HTH) domain after the proteins HB1, ASLX, and Restriction Endonuclease, the proteins in which it was detected. The HARE-HTH leucine-rich helical domain is not only present in several eukaryotic chromatin proteins but also in many prokaryotic key factors such as restriction endonucleases and RNA polymerase complexes. There are three helices within the HTH unit that form a distinct type of “wing helix-turn-helix”. HARE-HTH displays a specific conservation pattern, and considering its highly conserved structure in various essential proteins in human, fish, plants, chlorophyte algae, and red algae, it has been presented as indispensable for multidomain proteins that are involved in chromatin-related functions. It was also postulated that some HARE-HTH proteins might identify specific DNA modifications such as 5-methyl-cytosine
[21]. HARE-HTH is associated with a DDT motif (explained below in 1–6) and either of the homeo- or WAC motifs.
In the BAZ family of proteins, HARE-HTH, DDT, and WAC motifs are present and are separated by low-complexity regions. This pattern is also true of other factors such as plant HB1 and chlorophyte HDZ1, where HARE-HTH is associated with the DDT and homeodomain instead of WAC. There are three HARE-HTH motifs on BAZ1B (
Figure 4), the second and third of which overlap with the previously described conserved BAZ1 and BAZ2 motifs. These motifs were named WHIM (WSTF, HB1, Itc1p, MBD9) motifs 1, 2, and 3. Acf1 interaction with SNF2 is mediated by a region containing a DDT and three WHIM motifs, and it is postulated that ISWI binding is a common feature of WHIM-containing proteins. The BAZ1B WHIM1, 2, and 3 motifs span residues 724–773, 899–936, and 991–1032, respectively. WHIM is significantly associated with the DDT motif, and together, they form a binding pocket for the SLIDE domain of ISWI (
Figure 6). Highly conserved residues in WHIM1 work cooperatively with DDT and WHIM2 and mediate interaction with ISWI. WHIM motifs coupled with DDT domain are postulated as “protein rulers” that can regulate nucleosome spacing
[21].
Figure 6. Schematic prediction of SNF2H SANT domain bound to DNA (PDB ID:6NE3) and BAZ1B protein (PDB ID: Q9UIG0) facing it with an ISWI binding pocket comprised of DDT, WHIM1, WHIM2, and WHIM3 domains.
7. DDT and LXXLL Motifs
The DDT domain, which is found in DNA-binding homeobox-containing proteins and different transcription and chromatin remodeling factors was first described by Doerks et al. through homology-based sequence analysis
[20]. It consists of ~60 residues with conserved N-terminal phenylalanines, several aromatic and charged residues, and C-terminal leucines. It contains three α helices and exclusively associates with common domains found in nuclear proteins. In addition to bromodomain PHD finger transcription factors (BPTF) and putative DNA-binding proteins, DDT has also been identified in the BAZ family.
A leucine-rich helix domain was also reported by Jones et al. and was called the LH domain, which is a conserved region and contains a Leu-Xaa-Xaa-Leu-Leu (LXXLL) motif in an almost identical position between the WAC and BAZ1 domains among all BAZ members. BAZ1B displays two tandemly arranged LXXLL motifs where BAZ1BL, which is a structural variant of BAZ1B because it has 12 additional nucleotides in exon 7, harbors an additional LXXLL motif next to the original two
[13]. The LXXLL motif was first reported in 1997 as a short, conserved motif that is capable of and sufficient to bind transcriptionally active nuclear receptors
[29]. These nuclear receptor box motifs have been implicated in several bromodomain proteins and are suggested to be responsible for binding to nuclear receptors for retinoic acid, estrogen, progesterone, and vitamin D3
[30]. For instance, there are three LXXLL motifs within the core interaction domain of human SRC-1a that mediate its binding to estrogen receptors (ER)
[29].
8. Other Motifs and Conserved Regions
A FERM domain has also been identified toward the middle of BAZ1B between the DDT and BAZ1 domains
[31] (
Figure 4). This domain has been assigned several different names, including the amino-terminal domain, membrane-cytoskeletal-linking domain, erzin-like domain of the band 4.1 superfamily, the ERM like domain, and the conserved N-terminal domain; however, FERM, which stands for
4.1 protein erzin radixin meosin, has subsequently been proposed as a consensus naming format
[32]. It contains multiple phosphorylation and N-linked glycosylation sites and is mainly composed of β-sheet structures in addition to a few α-helices. This hydrophobic cysteine-rich domain is involved in protein-mediated cytoskeleton attachment to the plasma membrane and in maintaining cell integrity and mobility
[33]. The presence of this domain in a nuclear protein is somewhat unexpected, and what function, if any, it performs in BAZ1B remains unclear.
Two PEST sequences were identified in BAZ1B
[2] that are common among proteins with a short intracellular half-life. Many PEST sequences are conditional proteolytic signals for rapid degradation that can be activated in several ways. These sequences are enriched in proline (P), glutamic acid (E), serine (S), and threonine (T) residues; are uninterrupted but confined by positively charged amino acids (lysine, arginine, or histidine); and are found in various functionally important proteins such as oncogenes (e.g., adenovirus early region 1A (E1A), c-fos, p53), transcription factors (e.g., c-myc, v-myb), key metabolic enzymes (e.g., ornithine decarboxylase (ODC)), cyclins, and protein phosphatases
[34][35]. A number of PEST domains have been implicated as anchor sites for the E3 ubiquitin ligases involved in unbiquitin-mediated protein degradation
[36]. Considering their presence in long-lived proteins too, alternative functions are also suggested for PEST domains including intracellular sorting, binding of the SUMO-conjugating enzyme Ubc9, and modulation of the inward-rectifier potassium channel
IK1 [37].