Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Sigalit Rachel Sukenik | + 1616 word(s) | 1616 | 2021-09-16 05:59:00 | | | |
| 2 | Rita Xu | + 121 word(s) | 1737 | 2021-09-30 03:29:13 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Sukenik, S. Arsenite Inducible Regulatory Particle-Associate Protein. Encyclopedia. Available online: https://encyclopedia.pub/entry/14740 (accessed on 08 February 2026).
Sukenik S. Arsenite Inducible Regulatory Particle-Associate Protein. Encyclopedia. Available at: https://encyclopedia.pub/entry/14740. Accessed February 08, 2026.
Sukenik, Sigalit. "Arsenite Inducible Regulatory Particle-Associate Protein" Encyclopedia, https://encyclopedia.pub/entry/14740 (accessed February 08, 2026).
Sukenik, S. (2021, September 29). Arsenite Inducible Regulatory Particle-Associate Protein. In Encyclopedia. https://encyclopedia.pub/entry/14740
Sukenik, Sigalit. "Arsenite Inducible Regulatory Particle-Associate Protein." Encyclopedia. Web. 29 September, 2021.
Copy Citation
Maintaining 26S proteasome activity under diverse physiological conditions is a fundamental requirement in order to maintain cellular proteostasis. Several quantitative and qualitative mechanisms have evolved to ensure that ubiquitin–proteasome system (UPS) substrates do not accumulate and lead to promiscuous protein–protein interactions that, in turn, lead to cellular malfunction.
AIRAP
PSMD14/RPN11
proteasome
protein misfolding
1. Introduction
Maintaining proteome functionality is a major challenge that the cell must resolve in order to survive the diverse conditions that it encounters. Changes in biophysical conditions and post-translational modifications all impact the correct folding of proteins, and cellular machineries have evolved to maintain proteome integrity. Promiscuous interactions of misfolded proteins are highly toxic to the cell and manifest in various pathologies [1]; therefore, restoring the proper configuration of misfolded proteins by chaperones, reducing enzymes and more constitutes a salvation pathway. In contrast, degradation pathways such as the ubiquitin–proteasome system (UPS) or the lysosomal pathway eliminate the toxicity of misfolded proteins by proteolytic degradation [2]. The 26S proteasome is the central module of the UPS to which all designated proteins, usually ubiquitinated with suitable configurations of ubiquitin, are delivered. The 26S proteasome is composed of a 20S catalytic particle and a 19S regulatory particle that enables recognition, deubiquitination and unfolding of a UPS substrate. In order to ensure the functionality of the basic 26S proteasome module, various modes of regulation exist in order to maintain functionality during acute and chronic conditions. We can artificially divide these modifications into several categories, including quantitative and qualitative changes in proteasome subunits (executed by transcriptional [3][4][5] and translational [6] programs as well as post-translational modifications of proteasome subunits [7]) and changes in the composition of specific subunits, as exemplified in the case of the immunoproteasome (20Si) and the thymus proteasome (20St). In this mode of regulation, the exchange of catalytic subunits in response to acute signaling (IFN-γ) [8][9] or unique surroundings [10][11] enables modification of the proteasome to a more suitable peptide output. Another mode of regulation consists of alternating the entire cap composition. Alternative caps such as PA200 enable the engagement of the proteasome with specific substrates [12][13], while the PA28 (11S) enables proteasome modulation during specific conditions such as IFN-γ [9][14][15]. The fourth mode of regulation that is diverse and continuously engaged with the proteasome involves shuttling factors. These accessory factors typically consist of a proteasome binding domain that usually consists of a ubiquitin-like (UBL) domain that binds one of the proteasome ubiquitin receptors and several ubiquitin binding domains (UBD) that account for ubiquitin chain affinity and specificity (for reviews, see [16][17][18]). The UBA–UBL mode of substrate delivery to the proteasome increases the diversity and flux of UPS substrates [19], induces proteasome activation [20] and supports the formation of liquid–liquid phase separation of proteasomes [21][22].
Originally identified as an arsenite inducible gene [23], AIRAP/Zfand2a was subsequently found to bind proteasomes with unique biochemical characteristics [24]. These include AIRAP–proteasome integrity in the absence of ATP and enhanced peptide hydrolysis. Unlike other proteasomal adaptors, AIRAP–proteasome binding is not obtained via UBA domains, nor does it bind ubiquitin. AIRAP knockout cells accumulate higher polyubiquitin levels in response to arsenite exposure (typical of UPS impairment) and proteasomes acquired from the knockout cells contain higher levels of polyubiquitinated substrates [24]. These findings rule out AIRAP’s function as a shuttle factor but rather indicate it as a proteasome adaptor required for the efficient processing of UPS substrates by the proteasome upon arsenite-inflicted inhibition. In line with this notion, AIRAP expression was found to be induced upon proteasomal inhibition [25]; thus, AIRAP’s role and gene expression programing point towards a mechanistic function in retaining proteasomal function upon inhibitory conditions, yet the mechanistic details of AIRAP function have not been revealed.
2. Recombinant AIRAP Binding to Arsenite
Arsenite-induced protein misfolding occurs through binding to vicinal cysteine residues [26]. AIRAP is a short polypeptide highly enriched with cysteine residues that coordinate through two AN1 type zinc fingers the binding of zinc residues. To evaluate the possibility of the direct binding of AIRAP to arsenite, we utilized an immobilized bivalent form of arsenite (phenylarsine oxide (PAO)) previously demonstrated to bind arsenite-binding proteins [27].
Recombinant AIRAP was incubated with PAO beads, unbound material was extensively washed, and AIRAP immunoblot was performed to evaluate the bound content (Figure 1). Several controls were performed to evaluate the nature of this binding, and the protein configuration was evaluated by preincubating the recombinant protein with guanidine. The modification of AIRAP cysteine content was evaluated by preincubation with N-ethylmaleamide (NEM) or the reducing agent THP. Finally, a trivalent arsenite (As3+) and a bivalent form of arsenite were used to evaluate their ability to outcompete the immobilized PAO binding to AIRAP. As evident from the results in Figure 1A, the binding of AIRAP was highly dependent on the protein’s native structure (guanidine) and cysteine configuration (NEM and THP). Furthermore, AIRAP binding efficiently competed with a trivalent form of arsenite but not with a bivalent form (arsenite and PAO). To further investigate the specificity of AIRAP’s ability to bind arsenite and the effect of different metals on AIRAP’s ability to bind the PAO matrix, we incubated recombinant AIRAP with PAO matrix followed by incubation with various metals. As indicated in Figure 1B, metals such as As3+ and Fe3+ reduced AIRAP’s binding to the PAO matrix (Figure 1B, lanes 3 and 7). These results imply a selective metal binding by AIRAP to the PAO matrix.
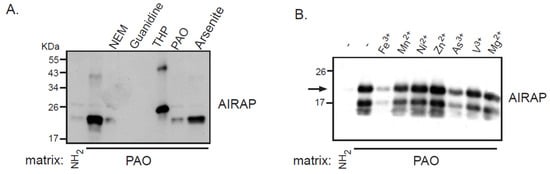
Figure 1. Recombinant AIRAP binding to a PAO matrix. (A) Recombinant mouse AIRAP was expressed as a C-terminus 6×His fusion protein and Ni-NTA affinity purified. Purified material was incubated with a control (NH2) or amino-phenylarsine (PAO) immobilized Sepharose 4B matrix in the presence of the cysteine alkylating N-ethylmaleamide (NEM), denaturing conditions (6M guanidine–HCl), reducing conditions (THP), sodium arsenite (As) or soluble amino-phenylarsine (PAO). Immobilized material was extensively washed, eluted by boiling with 2% SDS and subjected to SDS-PAGE and subsequent immunobloting to evaluate AIRAP content. (B) The impact of metals on AIRAP binding to a PAO matrix was performed as in 1A by performing the binding in the presence of the indicated metals. The arrow indicates the full-length recombinant product of AIRAP.
As AIRAP is a zinc-binding protein, zinc may serve as a primary structural enhancer to enable the optimal positioning of cysteine residues to bind arsenite. This notion is further supported by our results with the PAO matrix (Figure 1B). To directly measure the protein’s capacity to bind arsenite in a zinc-dependent manner, we measured arsenite binding to AIRAP by inductively coupled plasma optical emission spectroscopy (ICP-OES). Purified AIRAP was incubated with increasing concentrations of arsenite in the presence or absence of zinc, and arsenite content was evaluated. As seen in Figure 2A, AIRAP binding to arsenite was greatly enhanced when first incubated with zinc (marked Zn+). These results coincided with the indicated increase in AIRAP binding to the PAO matrix upon zinc incubation (Figure 1B). Analytical evaluation of this interaction was performed to determine the Kd value of AIRAP to arsenite using a micro-scale thermophoresis (MST) methodology [28]. A Kd of 7 nM ± 0.87 nM was determined (Figure 2B), demonstrating the high affinity of AIRAP toward arsenite.
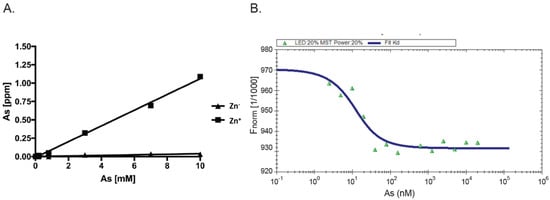
Figure 2. Dependence on zinc and MST measured affinity. (A) The presence of arsenite bound to AIRAP was determined by ICP-OES. Recombinant AIRAP was incubated in the presence (Zn+) or absence (Zn−) of zinc, prior to arsenite exposure. Bound material was extensively washed and arsenite content determined. (B) Recombinant AIRAP (1 μM) was incubated with a range of arsenite dilutions as indicated, and intrinsic fluorescence was measured by MST. Analysis revealed Kd values of 7 nM ± 0.87 nM.
3. AIRAP Binding to Arsenite In Vivo
After examining the in vitro binding of AIRAP to arsenite, we addressed the ability of endogenous AIRAP to bind arsenite. To this end, we used the cell lysate as a source for AIRAP. Upon endogenous expression of AIRAP (induced by arsenite treatment), AIRAP could not be detected on the PAO matrix from total lysate (Figure 3, lanes 2 and 3) but could be detected from purified proteasomes (Figure 3, lanes 5 and 6). It is important to note that the interaction was stable even after a high-salt wash, as would be expected from a metal-coordinated binding to the PAO matrix (lane 6). In addition, although catalytic particle subunit PSMA1 was present in the total lysate and the proteasome fractions (Figure 3 lane 4), it was barely detectable in the PAO matrix-purified fractions; thus, PSMA1 served as a specificity control for the PAO matrix binding. We reasoned that the lack of AIRAP binding to the PAO matrix in the total lysate may have been due to the existence of other proteins that competed towards the PAO matrix binding, or it may have indicated a structural change that AIRAP may undergo upon proteasome binding. This transition may be intended towards optimal binding to arsenite only upon proteasome binding.
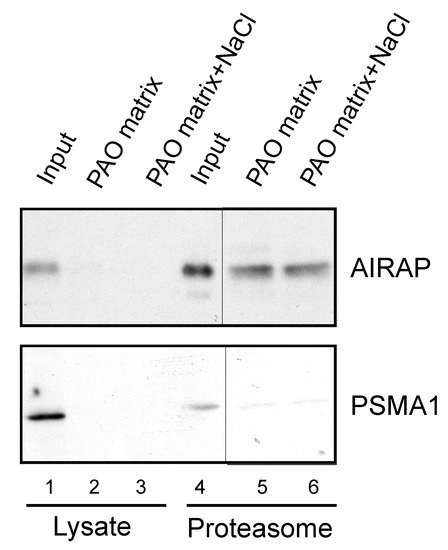
Figure 3. Endogenous proteasomal AIRAP binds arsenite. Cell lysates acquired from arsenite-treated cells were incubated with a PAO matrix. AIRAP and 20S proteasomal content (PSMA1) were evaluated by immunoblot (lanes 1–3). Alternatively, PAO matrix binding was performed using soluble proteasomes purified from arsenite-treated cells (lanes 4–6). Where indicated, PAO matrix was washed with high salt to evaluate impact on PAO binding (lanes 3 and 6).
4. Endogenous Rpn11/PSMD14 Binds PAO Matrix More Strongly Than Other Proteasome Subunits
After establishing the ability of AIRAP to bind arsenite, we sought to understand the extent to which arsenite-binding proteins bind the proteasome and how this function may relieve UPS impairment upon arsenite exposure. We reasoned that the 26S proteasome has one metal-binding protein, Psmd14/Rpn11, that functions as a proteasomal deubiquitination enzyme [29][30]. Possible impairment of Psmd14 activity may occur if arsenite binding perturbs metalloprotease activity, as previously reported for Rpn11 metal selectivity [31]. This, in turn, will lead to polyubiquitin accumulation and impaired turnover of UPS substrates. We therefore sought to determine whether Psmd14 can bind the PAO matrix. Affinity-purified soluble proteasomes were mixed with the PAO matrix and the Psmd14, S5a (an additional 19S subunit) and Psma1 (20S subunit) content was evaluated by immunoblot. As seen in Figure 4, Psmd14 was enriched on the PAO matrix. It is important to note that other proteasome subunits, while present, were not enriched as significantly as Psmd14 on the PAO matrix (compare PAO matrix lane and input lane for all evaluated proteasome subunits). These results are in line with Psmd14 being the proteasomal target for AIRAP binding.
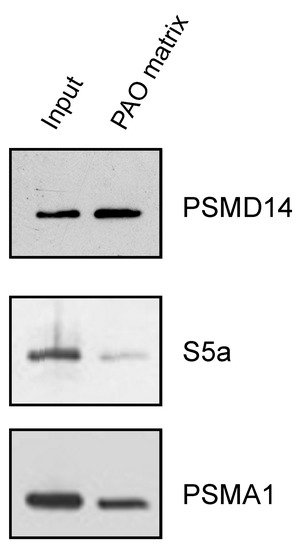
Figure 4. PSMD14 binding to PAO. Soluble proteasomes were incubated with PAO matrix, extensively washed and PSMD14, 19S (S5a), and 20S (PSMA1) content revealed by immunoblot.
References
- Bucciantini, M.; Giannoni, E.; Chiti, F.; Baroni, F.; Formigli, L.; Zurdo, J.; Taddei, N.; Ramponi, G.; Dobson, C.M.; Stefani, M. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature 2002, 416, 507–511.
- Schmidt, M.; Finley, D. Regulation of proteasome activity in health and disease. Biochim. Biophys. Acta. 2014, 1843, 13–25.
- Mannhaupt, G.; Schnall, R.; Karpov, V.; Vetter, I.; Feldmann, H. Rpn4p acts as a transcription factor by binding to PACE, a nonamer box found upstream of 26S proteasomal and other genes in yeast. FEBS Lett. 1999, 450, 27–34.
- Radhakrishnan, S.K.; Lee, C.S.; Young, P.; Beskow, A.; Chan, J.Y.; Deshaies, R.J. Transcription factor Nrf1 mediates the proteasome recovery pathway after proteasome inhibition in mammalian cells. Mol. Cell 2010, 38, 17–28.
- Steffen, J.; Seeger, M.; Koch, A.; Krüger, E. Proteasomal degradation is transcriptionally controlled by TCF11 via an ERAD-dependent feedback loop. Mol. Cell 2010, 40, 147–158.
- Panasenko, O.O.; Somasekharan, S.P.; Villanyi, Z.; Zagatti, M.; Bezrukov, F.; Rashpa, R.; Cornut, J.; Iqbal, J.; Longis, M.; Carl, S.H.; et al. Co-translational assembly of proteasome subunits in NOT1-containing assemblysomes. Nat. Struct Mol. Biol. 2019, 26, 110–120.
- Hirano, H.; Kimura, Y.; Kimura, A. Biological significance of co- and post-translational modifications of the yeast 26S proteasome. J. Proteom. 2016, 134, 37–46.
- Ferrington, D.A.; Gregerson, D.S. Immunoproteasomes: Structure, function, and antigen presentation. Prog. Mol. Biol. Transl. Sci. 2012, 109, 75–112.
- Aki, M.; Shimbara, N.; Takashina, M.; Akiyama, K.; Kagawa, S.; Tamura, T.; Tanahashi, N.; Yoshimura, T.; Tanaka, K.; Ichihara, A. Interferon-gamma induces different subunit organizations and functional diversity of proteasomes. J. Biochem. 1994, 115, 257–269.
- Sasaki, K.; Takada, K.; Ohte, Y.; Kondo, H.; Sorimachi, H.; Tanaka, K.; Takahama, Y.; Murata, S. Thymoproteasomes produce unique peptide motifs for positive selection of CD8(+) T cells. Nat. Commun. 2015, 6, 7484.
- Frantzeskakis, M.; Takahama, Y.; Ohigashi, I. The Role of Proteasomes in the Thymus. Front. Immunol. 2021, 12, 646209.
- Qian, M.X.; Pang, Y.; Liu, C.H.; Haratake, K.; Du, B.Y.; Ji, D.Y.; Wang, G.F.; Zhu, Q.Q.; Song, W.; Yu, Y.; et al. Acetylation-mediated proteasomal degradation of core histones during DNA repair and spermatogenesis. Cell 2013, 153, 1012–1024.
- Ustrell, V.; Hoffman, L.; Pratt, G.; Rechsteiner, M. PA200, a nuclear proteasome activator involved in DNA repair. EMBO J. 2002, 21, 3516–3525.
- Raule, M.; Cerruti, F.; Benaroudj, N.; Migotti, R.; Kikuchi, J.; Bachi, A.; Navon, A.; Dittmar, G.; Cascio, P. PA28αβ reduces size and increases hydrophilicity of 20S immunoproteasome peptide products. Chem. Biol. 2014, 21, 470–480.
- Tanahashi, N.; Yokota, K.Y.; Ahn, J.Y.; Chung, C.H.; Fujiwara, T.; Takahashi, E.I.; George, N.; DeMartino, G.N.; Slaughter, C.A.; Toyonaga, T.; et al. Molecular properties of the proteasome activator PA28 family proteins and gamma-interferon regulation. Genes Cells 1997, 2, 195–211.
- Saeki, Y. Ubiquitin recognition by the proteasome. J. Biochem. 2017, 161, 113–124.
- Chen, X.; Htet, Z.M.; López-Alfonzo, E.; Martin, A.; Walters, K.J. Proteasome interaction with ubiquitinated substrates: From mechanisms to therapies. FEBS J. 2020.
- Finley, D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 2009, 78, 477–513.
- Chen, X.; Randles, L.; Shi, K.; Tarasov, S.G.; Aihara, H.; Walters, K.J. Structures of Rpn1 T1:Rad23 and hRpn13:hPLIC2 Reveal Distinct Binding Mechanisms between Substrate Receptors and Shuttle Factors of the Proteasome. Structure 2016, 24, 1257–1270.
- Collins, G.A.; Goldberg, A.L. Proteins containing ubiquitin-like (Ubl) domains not only bind to 26S proteasomes but also induce their activation. Proc. Natl. Acad. Sci. USA 2020, 117, 4664–4674.
- Yasuda, S.; Tsuchiya, H.; Kaiho, A.; Guo, Q.; Ikeuchi, K.; Endo, A.; Arai, N.; Ohtake, F.; Murata, S.; Inada, T.; et al. Stress- and ubiquitylation-dependent phase separation of the proteasome. Nature 2020, 578, 296–300.
- Dao, T.P.; Kolaitis, R.M.; Kim, H.J.; O’Donovan, K.; Martyniak, B.; Colicino, E.; Hehnly, H.; Taylor, J.P.; Castañeda, C.A. Ubiquitin Modulates Liquid-Liquid Phase Separation of UBQLN2 via Disruption of Multivalent Interactions. Mol. Cell 2018, 69, 965–978.
- Sok, J.; Calfon, M.; Lu, J.; Lichtlen, P.; Clark, S.G.; Ron, D. Arsenite-inducible RNA-associated protein (AIRAP) protects cells from arsenite toxicity. Cell Stress Chaperones 2001, 6, 6–15.
- Stanhill, A.; Haynes, C.M.; Zhang, Y.; Min, G.; Steele, M.C.; Kalinina, J.; Martinez, E.; Pickart, C.M.; Kong, X.P.; Ron, D. An arsenite-inducible 19S regulatory particle-associated protein adapts proteasomes to proteotoxicity. Mol. Cell 2006, 23, 875–885.
- Rossi, A.; Riccio, A.; Coccia, M.; Trotta, E.; La Frazia, S.; Santoro, M.G. The proteasome inhibitor bortezomib is a potent inducer of zinc finger AN1-type domain 2a gene expression: Role of heat shock factor 1 (HSF1)-heat shock factor 2 (HSF2) heterocomplexes. J. Biol. Chem. 2014, 289, 12705–12715.
- Shen, S.; Li, X.F.; Cullen, W.R.; Weinfeld, M.; Le, X.C. Arsenic binding to proteins. Chem. Rev. 2013, 113, 7769–7792.
- He, X.; Ma, Q. NRF2 cysteine residues are critical for oxidant/electrophile-sensing, Kelch-like ECH-associated protein-1-dependent ubiquitination-proteasomal degradation, and transcription activation. Mol. Pharmacol. 2009, 76, 1265–1278.
- Wienken, C.J.; Baaske, P.; Rothbauer, U.; Braun, D.; Duhr, S. Protein-binding assays in biological liquids using microscale thermophoresis. Nat. Commun. 2010, 1, 100.
- Yao, T.; Cohen, R.E. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature 2002, 419, 403–407.
- Verma, R.; Aravind, L.; Oania, R.; McDonald, W.H.; Yates, J.R.; Koonin, E.V.; Deshaies, R.J. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science 2002, 298, 611–615.
- Fuchs, A.C.D.; Maldoner, L.; Wojtynek, M.; Hartmann, M.D.; Martin, J. Rpn11-mediated ubiquitin processing in an ancestral archaeal ubiquitination system. Nat. Commun. 2018, 9, 2696.
More
Information
Subjects:
Biochemistry & Molecular Biology
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
729
Revisions:
2 times
(View History)
Update Date:
30 Sep 2021
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




