
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Alexander E Vinogradov | + 3233 word(s) | 3233 | 2021-10-28 11:52:26 | | | |
| 2 | Beatrix Zheng | -755 word(s) | 2478 | 2021-11-03 11:14:13 | | | | |
| 3 | Beatrix Zheng | -1 word(s) | 2477 | 2021-11-03 11:17:43 | | | | |
| 4 | Beatrix Zheng | Meta information modification | 2477 | 2021-11-05 07:07:05 | | |
Video Upload Options
The growth of complexity in evolution is a most intriguing phenomenon. Using gene phylostratigraphy, we showed this growth (as reflected in regulatory mechanisms) in the human genome, tracing the path from prokaryotes to hominids. Generally, the different regulatory gene families expanded at different times, yet only up to the Euteleostomi (bony vertebrates). The only exception was the expansion of transcription factors (TF) in placentals; however, we argue that this was not related to increase in general complexity. Surprisingly, although TF originated in the Prokaryota while chromatin appeared only in the Eukaryota, the expansion of epigenetic factors predated the expansion of TF. Signaling receptors, tumor suppressors, oncogenes, and aging- and disease-associated genes (indicating vulnerabilities in terms of complex organization and strongly enrichment in regulatory genes) also expanded only up to the Euteleostomi. The complexity-related gene properties (protein size, number of alternative splicing mRNA, length of untranslated mRNA, number of biological processes per gene, number of disordered regions in a protein, and density of TF–TF interactions) rose in multicellular organisms and declined after the Euteleostomi, and possibly earlier. At the same time, the speed of protein sequence evolution sharply increased in the genes that originated after the Euteleostomi. Thus, several lines of evidence indicate that molecular mechanisms of complexity growth were changing with time, and in the phyletic lineage leading to humans, the most salient shift occurred after the basic vertebrate body plan was fixed with bony skeleton. The obtained results can be useful for evolutionary medicine.
1. Introduction
2. Analysis on Results
3. Current Insights
Summary of the Main Points
(1) Methodologically, we introduced the distinction between the ‘deep’ and ‘shallow’ phylostratigraphy, compared these approaches, and discussed them in the context of the classic ‘lumping vs. splitting’ problem (thereby extending it to the molecular field). This helps explain the controversial gene datings that were published previously.
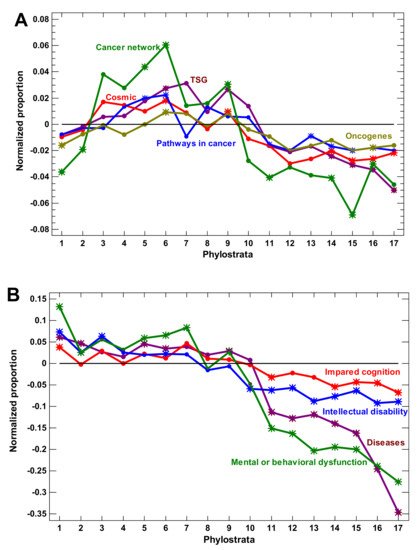
(2) Surprisingly, although transcription factors (TF) originated in the Prokaryota while chromatin appeared only in the Eukaryota, the expansion of epigenetic factors predated the expansion of TF. Protein modifiers and epigenetic factors expanded at the unicellular evolutionary stage, whereas TF and signaling receptors expanded in the multicellular organisms. Because cancer is an atavistic shift to unicellularity, these observations suggest that protein modifiers and epigenetic factors can be (at least) as important for oncogenesis as TF and signaling receptors.
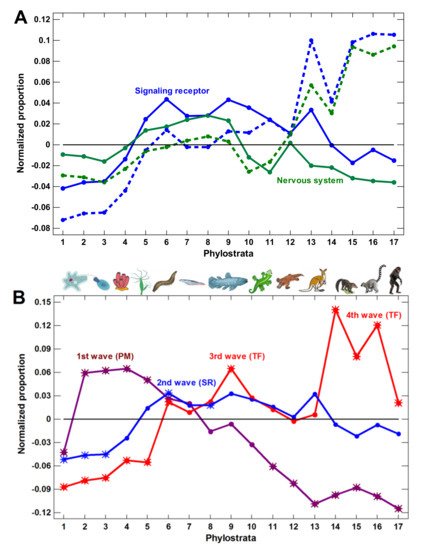
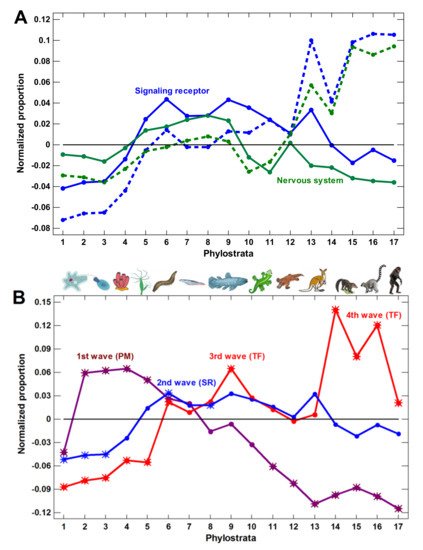
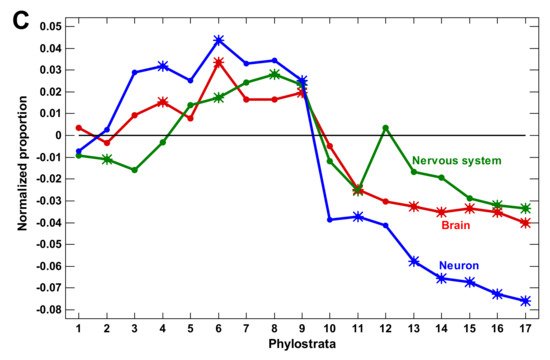
(3) The expansion of nervous system-related genes could create a misleading notion that the growth of complexity in the latest phylostrata was owing to these genes. However, this later expansion was due to olfactory receptors, which reflected only the elaboration of olfaction. Without olfactory receptors, the nervous system genes expanded only up to the Euteleostomi (bony vertebrates).
(4) Several lines of evidence suggest a salient shift in the evolution of complexity after the Euteleostomi. The expansions of regulatory gene families as well as the disease genes (indicating vulnerabilities in a complex organization and strongly enriched in regulatory genes) sharply declined after the Euteleostomi, with one paradoxical exception (explained below). The complexity-related gene properties (protein size, number of alternative splicing mRNA, length of untranslated mRNA, number of biological processes per gene, number of disordered regions in a protein, and density of TF–TF interactions) rose in multicellular organisms and declined after the Euteleostomi or earlier. At the same time, the speed of protein sequence evolution sharply increased.
(5) The above-mentioned exception is a unique expansion of TF families of deeply unicellular origin in the placentals, whereas the earlier expansion of TF was of purely multicellular origin. This inversion of TF origins and expansions creates a paradox, which can be explained as follows. It is likely that expansions of deeply unicellular TF occurred in the past, beginning from early times, but now we can see only their traces. They were situational expansions that occurred for the purpose of coping with invading transposons (‘nuclear immunity’) and deteriorated after the diminishing of the activity of cognate transposons. Only a minor part of these TF were adopted for other organism’s needs and retained (we showed the remnants of earlier expansions). Therefore, the recent expansion of deeply unicellular TF was related to clades rather than to grades of complexity. Thus, the expansions of olfactory and ‘nuclear immunity’ genes seem to be two possible pitfalls in the analysis of the impact of the gene repertoire on the growth of complexity.
4. Conclusions
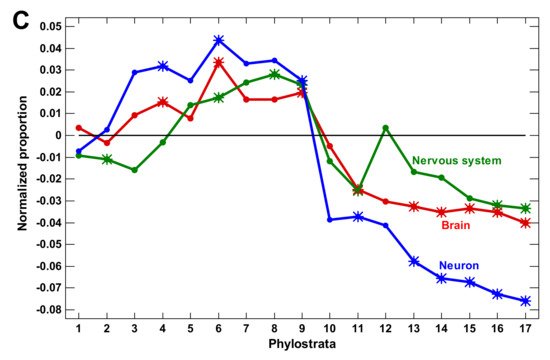
We concluded that the molecular mechanisms of complexity growth were changing with time, and after the basic vertebrate body plan was fixed with bony skeleton (in the Euteleostomi), there was a salient shift in these mechanisms within the phyletic lineage leading to humans. The first growth wave involved the expansion of protein modifiers (including epigenetic factors) that represented regulation at the level of gene products. They showed a plateau at the Eukaryota-Metazoa (unicellular sponges) and a depression after the Chordata. The second wave involved signaling receptors (representing intercellular communication), closely followed by a wave of transcription factors (probably representing diversification of cell types). These genes showed significant enrichment at the Bilateria and decline after the Euteleostomi. The wave of the nervous system genes generally coincided with the growth of other regulatory genes. This was probably because the nervous system should integrate all the other complexity waves at the organismal level. After the Euteleostomi, we revealed no significant expansions in gene groups that were related to general complexity (albeit there was an elaboration of olfaction and ‘nuclear immunity’). The overall picture of the main changes in the regulatory gene repertoire is shown in Figure 3.
The post-Euteleostomi complexity growth probably proceeded mostly via changes in protein sequences (the speed of which sharply increased) and in gene expression, with small-scale changes in the gene repertoire. The changes in gene expression could have been affected by a multitude of means, including changes in cis-regulatory elements, non-coding RNA, the structure of chromatin, and complex interactions of TF between themselves and with TF cofactors and epigenetic factors. The changes in TF complexes could have affected both transcription initiation and promoter-proximal pausing [34]. The invasion and propagation of transposable elements could also have changed the gene expression patterns [35][36]. Furthermore, there could have arisen intricate combinations of variation in terms of non-coding DNA, chromatin structure, TF, and epigenetic factors (with a growing space of splicing variants), followed by elaborate (post-) translational regulation.
Beginning from a certain threshold number of the basic genetic elements (protein-coding genes), the combinatorial space of protein products of these elements may have become large enough such that the further growth of organismal complexity could proceed without significant expansion in the genetic elements’ base. Natural selection explores this combinatorial space via changes in gene expression and protein interactions. Finally, the expansion of regulation to the higher organization level (from regulatory genes to regulatory cells) seemed to be involved, which was manifested in the increase in the number of neurons (approximated by encephalization quotient).
All these possibilities cannot so far be traced, in the evolutionary path leading to humans, in a consistent and quantitative way that is similar to that of protein-coding sequences. Therefore, we should include a caveat that our approach embraces only a part of the possible mechanisms of complexity growth. In the future, other aspects can be added to this picture.
References
- Gould, S.J. The Structure of Evolutionary Theory; Harvard University Press: Cambridge, MA, USA, 2002; ISBN 978-0-674-41792-2.
- Torres-Paz, J.; Hyacinthe, C.; Pierre, C.; Rétaux, S. Towards an integrated approach to understand Mexican cavefish evolution. Lett.2018, 14, 20180101, doi:10.1098/rsbl.2018.0101.
- Vinogradov, A.; Anatskaya, O. Gene Golden Age paradox and its partial solution. Genomics2019, 111, 115–126, doi:10.1016/j.ygeno.2018.02.015.
- Sasakura, Y.; Hozumi, A. Formation of Adult Organs through Metamorphosis in Ascidians. Wiley Interdiscip. Rev. Dev. Biol.2018, 7, e304, doi:10.1002/wdev.304.
- Guex, J. Retrograde Evolution During Major Extinction Crises; SpringerBriefs in Evolutionary Biology; Springer International Publishing: Cham, Switzerland, 2016; ISBN 978-3-319-27916-9.
- Lineweaver, C.H.; Davies, P.C.W.; Ruse, M. (Eds.) Complexity and the Arrow of Time; Cambridge University Press: Cambridge, UK,2013; ISBN 978-1-139-22570-0.
- Bains, W.; Schulze-Makuch, D. The Cosmic Zoo: The (Near) Inevitability of the Evolution of Complex, Macroscopic Life. Life2016, 6, 25, https://doi.org/10.3390/life6030025.
- Bizzarri, M.; Naimark, O.; Nieto-Villar, J.; Fedeli, V.; Giuliani, A. Complexity in Biological Organization: Deconstruction (and Subsequent Restating) of Key Concepts. Entropy2020, 22, 885, https://doi.org/10.3390/e22080885.
- Hahn, M.W.; Wray, G.A. The G-Value Paradox. Dev.2002, 4, 73–75, doi:10.1046/j.1525-142x.2002.01069.x.
- Vinogradov, A.E. Genome Size and Chromatin Condensation in Vertebrates. Chromosoma2005, 113, 362–369, doi:10.1007/s00412-004-0323-3.
- Taft, R.J.; Pheasant, M.; Mattick, J.S. The Relationship between Non-Protein-Coding DNA and Eukaryotic Complexity. Bioessays2007, 29, 288–299, doi:10.1002/bies.20544.
- Vinogradov, A.E.; Anatskaya, O.V. Organismal Complexity, Cell Differentiation and Gene Expression: Human over Mouse. Nucleic Acids Res.2007, 35, 6350–6356, doi:10.1093/nar/gkm723.
- Chen, C.-Y.; Chen, S.-T.; Juan, H.-F.; Huang, H.-C. Lengthening of 3’UTR Increases with Morphological Complexity in Animal Evolution. Bioinformatics2012, 28, 3178–3181, doi:10.1093/bioinformatics/bts623.
- Chen, L.; Bush, S.J.; Tovar-Corona, J.M.; Castillo-Morales, A.; Urrutia, A.O. Correcting for Differential Transcript Coverage Reveals a Strong Relationship between Alternative Splicing and Organism Complexity. Biol. Evol.2014, 31, 1402–1413, doi:10.1093/molbev/msu083.
- Dicke, U.; Roth, G. Neuronal factors determining high intelligence. Trans. R. Soc. B Biol. Sci.2016, 371, 20150180, https://doi.org/10.1098/rstb.2015.0180.
- Yruela, I.; Oldfield, C.J.; Niklas, K.J.; Dunker, A.K. Evidence for a Strong Correlation Between Transcription Factor Protein Disorder and Organismic Complexity. Genome Biol. Evol.2017, 9, 1248–1265, https://doi.org/10.1093/gbe/evx073.
- Niklas, K.J.; Dunker, A.K.; Yruela, I. The evolutionary origins of cell type diversification and the role of intrinsically disordered proteins. Exp. Bot.2018, 69, 1437–1446, https://doi.org/10.1093/jxb/erx493.
- Martin, A.; Orgogozo, V. The Loci of Repeated Evolution: A Catalog of Genetic Hotspots of Phenotypic Variation. Evolution2013, 67, 1235–1250, doi:10.1111/evo.12081.
- Loehlin, D.W.; Ames, J.R.; Vaccaro, K.; Carroll, S.B. A major role for noncoding regulatory mutations in the evolution of enzyme activity. Natl. Acad. Sci.USA 2019, 116, 12383–12389, https://doi.org/10.1073/pnas.1904071116.
- Davies, P.; Lineweaver, C. Cancer tumors as Metazoa 1.0: tapping genes of ancient ancestors. Biol.2011, 8, 015001–015001, https://doi.org/10.1088/1478-3975/8/1/015001.
- Vincent, M. Cancer: A de-Repression of a Default Survival Program Common to All Cells?: A Life-History Perspective on the Nature of Cancer. Bioessays2012, 34, 72–82, doi:10.1002/bies.201100049.
- Trigos, A.S.; Pearson, R.B.; Papenfuss, A.T.; Goode, D.L. Altered Interactions between Unicellular and Multicellular Genes Drive Hallmarks of Transformation in a Diverse Range of Solid Tumors. Natl. Acad. Sci. USA2017, 114, 6406–6411, doi:10.1073/pnas.1617743114.
- Bussey, K.J.; Cisneros, L.H.; Lineweaver, C.H.; Davies, P.C.W. Ancestral Gene Regulatory Networks Drive Cancer. Natl. Acad. Sci. USA2017, 114, 6160–6162, doi:10.1073/pnas.1706990114.
- Trigos, A.S.; Pearson, R.B.; Papenfuss, A.T.; Goode, D.L. Somatic Mutations in Early Metazoan Genes Disrupt Regulatory Links between Unicellular and Multicellular Genes in Cancer. Elife2019, 8, e40947, doi:10.7554/eLife.40947.
- Vinogradov, A.; Anatskaya, O. Evolutionary framework of the human interactome: Unicellular and multicellular giant clusters. Biosystems2019, 181, 82–87, https://doi.org/10.1016/j.biosystems.2019.05.004.
- Vinogradov, A.E.; Anatskaya, O.V. Cell‐cycle dependence of transcriptome gene modules: comparison of regression lines. FEBS J.2020, 287, 4427–4439, https://doi.org/10.1111/febs.15257.
- Kyriazis, M. Ageing Throughout History: The Evolution of Human Lifespan. Mol. Evol.2019, 88, 57–65, https://doi.org/10.1007/s00239-019-09896-2.
- Stearns, S.C. Frontiers in Molecular Evolutionary Medicine. Mol. Evol.2019, 88, 3–11, https://doi.org/10.1007/s00239-019-09893-5.
- Lineweaver, C.H.; Davies, P.C.W.; Vincent, M.D. Targeting cancer’s weaknesses (not its strengths): Therapeutic strategies suggested by the atavistic model. BioEssays2014, 36, 827–835, https://doi.org/10.1002/bies.201400070.
- Trigos, A.; Pearson, R.B.; Papenfuss, A.T.; Goode, D.L. How the evolution of multicellularity set the stage for cancer. J. Cancer2018, 118, 145–152, https://doi.org/10.1038/bjc.2017.398.
- Khyeam, S.; Lee, S.; Huang, G.N. Genetic, Epigenetic, and Post-Transcriptional Basis of Divergent Tissue Regenerative Capacities Among Vertebrates. Genet. 2021, 2, e10042, doi:10.1002/ggn2.10042.
- Nguyen, P.D.; de Bakker, D.E.M.; Bakkers, J. Cardiac Regenerative Capacity: An Evolutionary Afterthought? Mol. Life Sci. 2021, 78, 5107–5122, doi:10.1007/s00018-021-03831-9.
- Domazet-Loso, T.; Brajković, J.; Tautz, D. A Phylostratigraphy Approach to Uncover the Genomic History of Major Adaptations in Metazoan Lineages. Trends Genet.2007, 23, 533–539, doi:10.1016/j.tig.2007.08.014.
- Core, L.; Adelman, K. Promoter-Proximal Pausing of RNA Polymerase II: A Nexus of Gene Regulation. Genes Dev.2019, 33, 960–982, doi:10.1101/gad.325142.119.
- Drongitis, D.; Aniello, F.; Fucci, L.; Donizetti, A. Roles of Transposable Elements in the Different Layers of Gene Expression Regulation. J. Mol. Sci.2019, 20, 5755, doi:10.3390/ijms20225755.
- Maupetit-Mehouas, S.; Vaury, C. Transposon Reactivation in the Germline May Be Useful for Both Transposons and Their Host Genomes. Cells2020, 9, 1172, doi:10.3390/cells9051172.




