
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Chongying Wang | + 4505 word(s) | 4505 | 2021-01-04 08:44:53 | | | |
| 2 | Camila Xu | Meta information modification | 4505 | 2021-02-08 07:07:25 | | |
Video Upload Options
The NAC (NAM, ATAF1/2, and CUC2) family of proteins is one of the largest plant-specific transcription factor (TF) families and its members play varied roles in plant growth, development, and stress responses.
1. Introduction
Crops are constantly challenged by a variety of abiotic and biotic factors that have negative impacts on their growth, development and yields [1]. Biotic stressors, mainly bacteria, fungi and viruses, can cause diseases and impact agricultural crops, forestry plantations, and native plant communities [2]. To resist or tolerate these adverse external stresses, plants have evolved many effective stress response strategies, including changes in morphology, establishing defensive systems, and constructing physiological, biochemical and molecular regulatory networks.
Plants have two main branches of defense against various pathogens: PAMP (pathogen-associated molecular patterns)-triggered immunity (PTI) and effector-triggered immunity (ETI) [3]. PTI is a basal defense mechanism in plants that is triggered by the recognition of PAMPs through PRRs (pattern recognition receptors) at the plant cell surface [4]. ETI is an accelerated and amplified PTI response, which arises from the interactions between plant resistance proteins and pathogen effector proteins [5][6]. Upon being attacked by pathogens, plants will activate a series of complex molecular regulatory networks, such as reactive oxygen species (ROS) signaling [7], phytohormone signaling [8], changes in redox status [9], and inorganic ion fluxes [10], to prevent further pathogen invasion.
Much of the plant response to pathogens involves transcriptional reprogramming. Plants have established high-efficiency gene expression networks to regulate multiple specific stress responsive genes in a coordinated manner. Such regulation of the large-scale expression of genes requires a concerted function of different types of transcription factors (TFs). TFs specifically bind to cis-elements and/or trans-acting factors in the promoters of target genes and act as transcriptional activators or repressors. Several TF families, such as NAC (NAM, ATAF and CUC), MYB (myeloblastosis-related proteins), WRKY (WRKYGQK), bZIP (basic leucine zipper domain), bHLH (basic helixloop-helix), CAMTA (CaM-binding transcription activator) and ERF/AP2 (ethylene responsive factor/apetala2), have crucial roles in abiotic and biotic stress responses [11]. So far, a large number of studies have proved that NAC TFs play roles in plant growth, development, abiotic stress response, and disease resistance [11][12][13][14][15][16][17].
Pathogens directly affect plant growth and development and decrease the quality and yields of the crop. Three major cereal crops, namely rice (Oryza sativa), maize (Zea mays), and wheat (Triticum aestivum), provide two-thirds of the food consumed all over the world [18]. Barley (Hordeum vulgare) and soybean (Glycine max) are also important crops, in the grain and legume families, while tomato (Solanum lycopersicum) is a fruit crop. NAC TFs comprise a gene family with 151 members in rice [19][20][21], 157 in maize [22], 559 in wheat [23], 167 in barley [24], 152 in soybean [25], and 93 in tomato [26]. Recent studies demonstrate that NAC TFs can be induced by pathogen infection, regulate the expression of downstream genes, and integrate hormone and other signals to offer plants resistance against pathogens.
2. Overview of NAC TFs
NAC TFs constitute one of the largest groups of plant-specific TFs, with more than 100 members in most plants, including model plants like Arabidopsis or most crops, and the highest reported number of 559 in wheat [23]. The NAC acronym is derived from three reported proteins that contain a highly conserved domain in their N-terminal region (the NAC domain): (i) NAM (No Apical Meristem), (ii) ATAF1/2 (Arabidopsis thaliana Transcription Activator Factor 1/2) and (iii) CUC2 (Cup-shaped Cotyledon 2) [20][21]. NAC TFs play essential roles in diverse biological processes, such as growth, development, senescence and morphogenesis, and are widely involved in signaling pathways in response to different phytohormones and multiple abiotic and biotic stress [27][28][29].
2.1. Phylogeny and Classification of the NAC TFs
NAC proteins are widespread in land plants, from “simple” bryophytes to “complex” angiosperms, and also in freshwater green algae (Charophytes) [30][31][32], a sister clade to land plants and likely their evolutionary root [33]. NAC proteins originated about 725–1200 million years ago (Mya), during the diversification of the charophytes/embryophytes (the Streptophytes), experienced a first expansion in bryophytes about 470 Mya, and a second expansion in angiosperms during the early Cretaceous period [27]. Along with the divergence of vascular plants, NAC proteins extensively expanded [30] shortly before the origination and radiation of angiosperms through different (segmental or tandem) duplications [31][34][35]. In angiosperms, there are two detectable evolutionary pathways of the NAC proteins: ancient duplications that occurred before the divergence of dicots and monocots and recent duplications in a particular lineage of dicots or monocots [30][36]. As a result of duplication, some family members possess redundant functions [37][38].
According to the clustering analyses by Zhu et al., NAC proteins consist of 21 subfamilies among bryophytes and vascular plants [30]. Among these subfamilies, 15 were found only in angiosperms, while the other six occur in both flowering plants and earlier diverged lycophytes [30][33]. In 2015, another team recategorized NAC proteins into six major orthologous groups (Group I-VI) using more than 2000 non-redundant sequences from 24 different species of green plants by comparative genomic and gene functional analyses [31]. NAC group I is considered the basal NAC group, because its members are involved in secondary wall and wood formation, which might have been essential to water conduction or support during the adaptation of plants to land environments [39][40]. Group II NACs function in specific developmental processes among different organs, as well as other processes such as ethylene-auxin pathways. NAC group III is named the TMM (transmembrane motifs) Group, as 142 of the 164 NAC proteins in this group contained a TMM in their C-terminal regions that can anchor them in biomembranes (especially on the endoplasmic reticulum), and can be hydrolyzed by proteolytic enzymes activated by signaling [41][42][43]. The members in NAC group IV have diverse functions, such as ANAC009, which controls the reorientation and timing of cell division, and ANAC042 which regulates longevity of Arabidopsis. NAC group V is proposed to be the Stress Group, as most of its members are involved in stress responses. Finally, NAC group VI contains many species-specific sequences, which experienced whole-genome duplication events through their evolutionary history [31].
2.2. Structure and Function of NAC Proteins
NAC proteins usually contain two relatively independent domains: one well-conserved N-terminal NAC domain of 151–159 amino acids and a relatively divergent C-terminal Transcriptional Activation Region (TAR) [20][44][45]. There are a few kinds of atypical NAC proteins, which have variations in either their NAC domains or TAR [46]. The NAC domain exists in all NAC proteins, and the most conserved consensus sequences are D-D/E-L-I/V, E-W-Y-F-F, G-Y-W-K, and M-H-E-Y [46]. The NAC domain contains nuclear localization signals (NLSs) and its main function is translocating the protein from the cytoplasmic matrix to the nucleus [44][47] and forming homo- or heterodimers, that the state in which these proteins bind DNA [48][49][50]. Some proteins also contain nuclear export signals (NESs) in the NAC domain, which might mediate their export out of the nucleus for degradation after their missions are completed [32]. Crystallographic structures of ANAC019 from Arabidopsis and SNAC1 (STRESS RESPONSIVE NAC1) from rice revealed that the NAC domain contains mainly twisted antiparallel β-sheet(s) flanked by a few α-helices. The central β-sheet is responsible for dimerization of the proteins, by forming stabilized salt bridges between conserved amino acids in two subunits, and also for interacting with the major groove of DNA in the dimer form [49][50][51]. The NAC domain can be further divided into five subdomains, designated A-E, distinguished by blocks of heterogeneous amino acids or gaps, among which subdomains A, C, and D are highly conserved compared to B and E. It is assumed that subdomain A promotes functional dimerization, subdomains C and D play parts in DNA binding, due to their content of basic amino acids, and in nuclear translocation of the protein with the help of the NLSs inside the subdomains, while the variable subdomains B and E diversify the roles of the NAC proteins [44][48].
The TAR region, also known as TRR (transcriptional regulatory region), is related to transcriptional regulation and corresponds to the distinct functions of NAC proteins [46][48], acting as both activator and/or repressor to downstream target genes together with the NAC domain [27]. Bioinformatic analysis revealed common motifs in the C-terminal regions of some NAC subfamilies [48][52]. In addition, an α-helical TMM could be found at the C-terminal end of many NAC proteins [43] that contain a conserved N-terminal NAC domain, a variable middle TAR region, and a C-terminal TMM motif [42]. With the help of the TMM, the nascent NAC protein can be anchored to the endoplasmic reticulum membrane or plasma membrane, retaining it outside of the nucleus in a dormant state. Subsequently, the NAC TF can be cleaved through either protease- or ubiquitin proteasome-mediated proteolytic events upon stimulation by internal and/or environmental signals, which would activate the mature nuclear-associated form to enter the nucleus and regulate downstream genes, which is an adaptive strategy to environment [41][42][43]. Recently, transmembrane domains were found in the N-terminal ends of a small number of NAC proteins [32].
3. NAC TFs Have Positive or Negative Roles in Crop Disease Resistance
In both model and crop plants, numerous NAC genes are induced in response to pathogen infection [46]. Over-expression or silencing of certain NAC genes results in enhanced or reduced resistance to pathogens [53][54][55][56], suggesting that NAC TFs could positively or negatively regulate plant defense response. Here, we introduce the roles of NAC TFs during the response to pathogens in several important crops, including rice, wheat, barley, potato, soybean, maize, tomato and lettuce.
3.1. Roles of NAC TFs in Rice
Rice is one of the most important food crops in the world, as it is consumed by more than 3 billion people [57]. Rice blast caused by the fungal pathogens Magnaporthe oryzae and M. grisea is one of the most devastating diseases threatening rice production. Several NACs have been found to respond to rice blast, therefore, it is important to understand the role of these TFs in response to both this disease and other pathogens in rice.
The rice genome is predicted to contain 151 NAC genes [19], but only a few have been characterized. OsNAC6 was the first NAC TF found to be involved in disease resistance in rice [58]. OsNAC6 can be induced by both abiotic and biotic stress, including blast disease. Transgenic rice plants over-expressing OsNAC6 had improved tolerance to rice blast, dehydration and high salt stress, but exhibited negative effects on growth and yields [58]. Similarly, resistance was obtained from the over-expression of OsNAC111 or OsNAC58. Over-expression of OsNAC111 or OsNAC58 in rice increased resistance to M. oryzae or the bacterial blight pathogen Xoo (Xanthomonas oryzae pv. oryzae), respectively [59][60]. In addition, inoculation with M. oryzae can induce OsNAC111 expression, which then activates several downstream defense genes, such as PR1 (PATHOGENESIS-RELATED1) and PR8, suggesting that the OsNAC111 TF positively regulates the expression of a specific set of PR genes in the rice blast response [59]. ONAC122 and ONAC131 also play positive regulatory roles during rice blast resistance and can be induced by infection with M. grisea and treatment with exogenous defense signaling molecules, such as salicylic acid (SA), methyl jasmonate (MeJA), or 1-aminocyclopropane-1-carboxylic acid (a precursor of ethylene) [61]. Metabolomics analysis was recently used, for the first time, to explore the molecular mechanism of NAC-mediated disease resistance. Liu et al. found that ONAC066 promotes the resistance against fungal blast and bacterial blight in rice by regulating the accumulation of soluble sugars and amino acids as well as the up-regulation of the PR gene [62].
The hypersensitive response (HR) of plants against disease is a type of programmed cell death. Kaneda et al. found that over-expression of OsNAC4 in rice could lead to HR cell death, accompanied with the loss of plasma membrane integrity, fragmentation of nuclear DNA, and typical morphological changes. HR cell death is noticeably decreased in OsNAC4 knock-down lines after induction by an avirulent pathogen N1141 [63], suggesting that OsNAC4 is a key positive regulator of plant hypersensitive cell death. In addition, the osnac60 mutant had increased susceptibility to M. oryzae, while the over-expressing plant had an enhanced defense response, including increased programmed cell death, ROS accumulation, and callous deposition and up-regulation of defense-related genes [64].
On the contrary, Yoshii et al. found that mutation of RICE DWARF VIRUS MULTIPLCATION 1 (RIM1), which encodes a NAC TF, resulted in a loss of susceptibility to Rice Dwarf Virus (RDV) but not to Rice Transitory Yellowing Virus (RTYV) and Rice Stripe Virus (RSV) [65]. The accumulation of virus capsid protein was significantly reduced in rim1 mutant plants inoculated with RDV, which impairs the multiplication of the virus, while proliferation of the virus was stimulated with over-expression of RIM1. Therefore, it was proposed that RIM1 negatively regulates rice resistance to RDV, by acting as a host factor that is required for multiplication of the virus [65].
This summary shows that 9 NAC TFs (OsNAC4, OsNAC6, OsNAC58, OsNAC60, ONAC066, OsNAC111, ONAC122, ONAC131, and RIM1) have been validated to take part in defense responses against pathogen attack. OsNAC6, OsNAC58, OsNAC60, ONAC066, OsNAC111, ONAC122 and ONAC131 positively regulate rice resistance to fungus Magnaporthe, while RIM1 negatively regulates rice resistance to the virus RDV.
3.2. Roles of NAC TFs in Wheat
Wheat is the most widely distributed food crop and occupies the largest planting area in the world [66]. The planting area and total output account for one third of all food crops, and about one-third of the population of the world uses wheat as its main food [67]. Thus, it is quite urgent to develop novel wheat varieties that have improved yield potential and increased tolerances to biotic and abiotic stresses. There are 559 TaNAC genes in the wheat genome, 186 of which were recently identified during infection of the resistant hexaploid wheat line N9134 with the fungal agents that cause rust and powdery mildew [23].
Powdery mildew, caused by the biotrophic Blumeria graminis f. sp. tritici (Bgt), is a widespread disease in wheat [68]. Zhou et al. isolated and characterized three wheat TaNAC6s, TaNAC6-A, TaNAC6-B and TaNAC6-D. Analysis of the expression patterns of the TaNAC6s showed that TaNAC6-A and TaNAC6-D were up-regulated early after Bgt inoculation. Over-expression of the TaNAC6s enhanced the resistance to Bgt, while silencing them reduced the resistance in wheat, indicating that the TaNAC6s play positive roles in resistance against Bgt [69]. Recently, wheat TaNACL-D1 was demonstrated to be involved in the disease response to Fusarium head blight (FHB) [70], a devastating disease of wheat and barley in humid and semi-humid regions of the world. TaNACL-D1 was recently shown to interact with TRITICUM AESTIVUM FUSARIUM RESISTANCE ORPHAN GENE (TaFROG), which enhances wheat resistance against FHB, and lines over-expressing TaNACL-D1 were more resistant to FHB disease [70][71].
Previous studies have shown that wheat TaNAC4 and TaNAC8 shared high homology with rice OsNAC4 and OsNAC8. TaNAC4 and TaNAC8 expression is induced by infection with Puccinia striiformis f. sp. tritici (Pst, which causes stripe rust) as well as by treatments with MeJA or ethylene (ET), but not by treatments with abscisic acid (ABA) or SA [72][73]. This indicates that TaNAC4 and TaNAC8 of wheat might be two important components in defense-signaling pathway and play essential roles in resistance to pathogen.
TaNAC1 is strongly expressed in wheat roots and is involved in responses to Pst infection and treatments with defense-related hormones. Knockdown of TaNAC1 enhances resistance to Puccinia stripe rust in wheat, while over-expression of TaNAC1 in Arabidopsis enhances susceptibility and reduces systemic-acquired resistance to Pseudomonas syringae pv tomato DC3000 (Pst DC3000). Similarly, knockdown of TaNAC21/22 in wheat enhanced resistance against Puccinia stripe rust [74][75]. Silencing of TaNAC2 or TaNAC30 enhances the resistance against stripe rust by significantly increasing H2O2 generation and decreasing hyphal growth at the early stage of the interaction between Pst and wheat [76][77]. These data indicate that TaNAC1, TaNAC2, TaNAC21/22, and TaNAC30 negatively regulate stripe rust resistance in wheat.
3.3. Roles of NAC TFs in Barley
Barley is the fourth most important grain crop in the world [78] and, like the other cereal crops, is threatened by various pests and diseases [79]. A search of the public barley sequence database identified 48 NAC genes (HvNACs), while the expression profiles of 46 HvNACs were investigated in various tissues with and without ABA or MeJA treatment. The HvNAC proteins have conserved functions in secondary cell wall biosynthesis, leaf senescence, root development, seed development and hormone-regulated stress response [80].
Barley HvNAC6 has a high similarity to the rice OsNAC6 in pathogen resistance. Transient over-expression of the gene in barley increased the penetration resistance of epidermal cells to the powdery mildew pathogen Blumeria graminis f. sp. hordei (Bgh), while silencing of the gene, using RNA interference (RNAi), enhanced the sensitivity to Bgh. Silencing of HvNAC6 also changed the accumulation of ABA, which was not affected by Bgh inoculation, indicating that HvNAC6 acts as an ABA-mediated defense response regulator to maintain basal resistance against Bgh [81][82]. In Arabidopsis, the expression of the HvNAC6 homologue ATAF1 was induced by Bgh, and the ataf1-1 mutant line displayed reduced penetration resistance to Bgh. HvNAC6 and ATAF1 play conserved positive roles in penetration resistance in both monocots and dicots, respectively [82].
Another NAC TF in barley is HvSNAC1, which promotes resistance to barley ramularia leaf spot (RLS) disease [83]. RLS is a new emerged barley disease, caused by the ascomycete fungus Ramularia collo-cygni and which broke out in Europe a decade ago [84]. Over-expression of HvSNAC1 in barley significantly reduced the severity of RLS, but had no effects on other pathogenic diseases, such as eyespot, powdery mildew, or blast. Further analysis showed that dark-induced leaf senescence is delayed in HvSNAC1 over-expression lines, indicating that HvSNAC1 may inhibit plant senescence [83].
3.4. Roles of NAC TFs in Tomato and Potato
Tomato is the highest value vegetable and fruit crop worldwide, at an annual production of 100 million tons, and makes a huge nutritional contribution to the human diet [85][86][87]. At the same time, tomato is constantly attacked by various pathogens, causing huge losses in production [88][89][90]. So far, 93 putative NAC proteins were identified in tomato [26]. Similar to other crops, tomato NAC TFs play roles in abiotic and biotic stress responses, as well as the development of the plant [91][92][93][94][95].
Infection by pathogens induces the expression of SlNAC1 in tomato, but plays dual functions in resistance to different pathogens. SlNAC1 expression is specifically induced in tomato by the replication enhancer (REn) of Tomato leaf curl virus (TLCV) [96], and its over-expression increases the accumulation of viral DNA in infected cells, indicating that SlNAC1 play negative roles in resistance against TLCV. SlNAC1 is also induced by Pseudomonas infection, but plays reversed roles in defense signaling [56]. In Pseudomonas-infected plants, expression of SlNAC1 increased rapidly while degradation of the SlNAC1 protein was suppressed. Further research proved that SlNAC1 could be ubiquitinated by SINA3, a ubiquitin ligase, but the expression of SINA3 was decreased in infected plants. Thus, pathogen infection counteracts the degradation of the SlNAC1 protein. These data suggest that SlNAC1 plays a positive role in resistance to Pseudomonas infection [56][97].
The NAC protein, Solanum lycopersicum Stress-related NAC1 (SlSRN1), was identified in tomato by virus-induced gene silencing technology [98]. The expression of SlSRN1 can be significantly induced by infection with Botrytis cinerea and Pst DC3000, while silencing of SlSRN1 leads to increased severity of the diseases. Silencing of SlSRN1 accelerates accumulation of ROS but reduces expression of defense genes after infection by B. cinerea. These results demonstrate that SlSRN1 is a positive regulator of the defense response against B. cinerea and Pst DC3000 in tomato [98]. Recently, six NAC TFs (SlNAC24, SlNAC20, SlNAC39, SlNAC47, SlNAC61 and SlNAC69) were studied in response to Tomato yellow leaf curl virus (TYLCV) infection in tomato. Four NAC genes (SlNAC20, SlNAC24, SlNAC47, and SlNAC61) were induced after TYLCV infection in resistant plants, and SlNAC61 played positive roles in response to TYLCV infection, according to Virus-induced gene silencing analysis. Furthermore, the six NAC TFs could interact with protein phosphatase 2C (PP2C), mitogen-activated protein kinase 3 (MPK3), and some defense response TFs, such as WRKY, MYB, and even NAC, by binding the promoters of these genes, indicating that NAC TFs have an complex response mechanism during TYLCV infection [94]. Recently, it was found that SlNAC082, a ribosomal stress mediator, was involved in the process of infection by citrus exocortis viroid (CEVd) in tomato. A higher expression level of SlNAC082 was detected in the CEVd-infected tomato leaves. CEVd and its derived viroid small RNAs were found to co-sediment with tomato ribosomes in vivo and caused alterations in ribosome biogenesis in the infected tomato plants. The alterations in both the rRNA processing and the induction of SlNAC082 were correlated with the degree of viroid symptomology [95].
Stomata play an active part in the plant innate immune response, and serve as an entrance for pathogen into plant cells [99]. The genes JA2 (Jasmonic Acid 2) and JA2L (JA2-like) both encode two NAC TFs that are closely related to ANAC019/ANAC055/ANAC072. These NACs were preferentially expressed in guard cells of tomato leaves [100]. In JA2-SRDX (SUPERMAN REPRESSION DOMAIN X) plants, Pst DC3000-induced stomatal closure was impaired at 1 h post infection (hpi), and pathogen-triggered stomatal reopening remained normal at 4 hpi, indicating that JA2 is required for Pst DC3000-induced stomatal closure but not stomatal reopening. By contrast, the Pst DC3000-induced stomatal closure was largely normal at 1 hpi, but the pathogen-triggered stomatal reopening was substantially impaired at 4 hpi in JA2L-AS plants expressing an antisense version of the JA2L cDNA, indicating that JA2L is required for pathogen-regulated stomatal reopening [100].
Potato is one of the four major food crops around the world. A NAC TF, StNACb4 from potato, was identified and characterised. StNACb4 has been shown to promote resistance to bacterial wilt caused by Ralstonia solanacearum [101]. Transgenic tobacco plants were generated in which the expression of StNACb4 was constitutively up-regulated or suppressed using RNAi. StNACb4 was found specifically in the phloem of the vascular system of the stems and leaves, and up-regulated upon infection with R. solanacearum or by treatment with SA, ABA and MeJA in transgenic tobacco. Silencing StNACb4 reduced the tolerance of tobacco to R. solanacearum, and over-expression of the gene enhanced the tolerance to this pathogen [101]. These results are consistent with findings on StNAC43, another potato NAC TF, which can increase the deposition of resistance-related metabolites to reinforce the secondary cell wall and improve resistance to late blight disease [102]. These data demonstrate that both StNACb4 and StNAC43 are positive regulators of disease resistance of potato.
3.5. Roles of NAC TFs in Other Crops
Maize is not only an important and widely distributed cereal crop, but also a model plant for genetic research [103]. Plant diseases induced by pathogens cause huge yield losses, up to 41.1% every year [104]. Lu et al. identified 157 non-redundant maize NAC genes, which were unevenly distributed on 10 maize chromosomes [22]. Further sequence and evolutionary relationship analysis showed that 19 maize NAC genes were related to stress responses [105]. ZmNAC41 and ZmNAC100 were transcriptionally induced during infection by Colletotrichum graminicola and defense signals, and were also expressed during leaf senescence in maize. In addition, ZmNAC41 was up-regulated in response to the fungal biotroph Ustilago maydis. Interestingly, the transcripts of ZmNAC41 and ZmNAC100 are induced by JA and SA, respectively, suggesting that ZmNAC41 and ZmNAC100 could function in the defense response [106]. When the upstream promoters of maize NAC genes were analyzed, a MYC binding site was detected in ZmNAC15, ZmNAC38 and ZmNAC41, while a WRKY-binding motif was detected in ZmNAC15, ZmNAC36, ZmNAC41, and ZmNAC100. In short, the ZmNAC15, ZmNAC36, ZmNAC38, ZmNAC41, and ZmNAC100 genes all contained potential binding elements for TFs known to be involved in the plant defense network [106].
Soybean is a main source of high-quality proteins and a vegetable oil that provide nutrition for animals and humans [107]. NAC TFs are believed to play vital roles in soybean development and disease resistance. Six NAC-like genes, designated GmNAC1–GmNAC6, were cloned and characterized from soybean a decade ago. These genes had similar genomic organization and high sequence similarity, especially in the NAC domains, but exhibited different expression patterns during seed development [108]. Subsequently, more NAC proteins were identified in soybean [109], most of which are involved in development and abiotic stress, such as GmNAC30 [110], GmNAC81 [110][111], GmNAC109 [112] and GmNAC8 [113], while up to now only GmNAC42 is reported to be involved in plant disease resistance [114][115][116]. Soybean GmNAC42-1 is a homolog of the Arabidopsis ANAC042-1, which is an indole alkaloid plant antitoxin regulator. Over-expression of GmNAC42-1 in elicited hairy roots significantly increases the amount of glyceollin in soybean, suggesting this protein is an essential and positive regulator of glyceollin biosynthesis. GmNAC42 is annotated as a systemic acquired resistance (SAR) gene and functions in soybean disease resistance because glyceollins are defensive metabolites (phytoalexins) derived from isoflavones in soybean [114]. MYB TFs are also involved in the glyceollin gene regulatory network. GmMYB29A1 and GmMYB29A2 were up-regulated in hairy roots treated with a wall glucan elicitor from P. sojae, and the expression of GmNAC42-1 and GmMYB29A1 were increased with the over-expression of GmMYB29A2, indicating that GmNAC42-1 was also regulated by GmMYB29A2 during glyceollin biosynthesis [116].
In lettuce, LsNAC069, a NAC TF with a C-terminal TMM motif, is a target of the RxLR-like effectors of the fungus Bremia lactucae [117]. RxLR effectors are characterized by a conserved RxLR (Arg-x-Leu-Arg) motif in the N-terminal domain, and B. lactucae secretes potential RxLR effectors during the infection process. LsNAC069 silencing increases resistance to Pseudomonas cichorii bacteria. LsNAC069 is relocalized from the ER to the nucleus when wild-type plants are treated with Phytophthora capsici culture filtrate, but this process could be prevented by the protease inhibitor TPCK (N-tosyl-L-phenylalanine chloromethyl ketone), indicating that the LsNAC069 needs proteolytic cleavage to be untethered from the ER and relocalized to the nucleus. However, the susceptibility to B. lactucae was not significantly altered in LsNAC069 silenced lettuce lines, and the process of LsNAC069 relocalization was inhibited upon the expression of B. lactucae effectors. Moreover, both co-localization and yeast two-hybrid experiments demonstrated that LsNAC069 could interact with B. lactucae effectors. Together these data demonstrate that B. lactucae can cause disease in lettuce through its RxLR effectors inhibiting the hydrolysis and relocalization of LsNAC069 from the ER to the nucleus, which suppresses the activation of genes downstream of LsNAC069 [117].
As mentioned above, we constructed a phylogenetic tree of NAC TFs cited in the review except LsNAC069 in lettuce, which cannot be searched out, and constructed a diagram of those genes (Figure 1). Meanwhile, we also made a diagram of the NAC domain structure of most NAC protein sequences mentioned here (Figure 2).
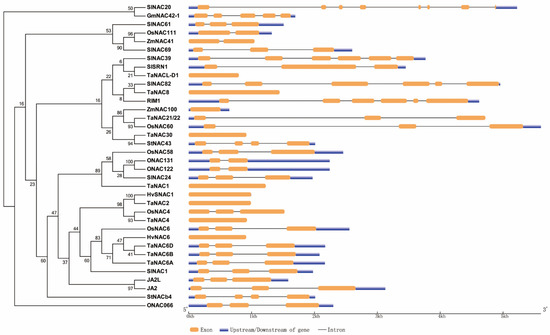
Figure 1. Phylogenetic and gene structure analysis of NAC TFs mentioned in this review. The left is phylogenetic analysis of some NAC TFs from rice, wheat, barley, maize, soybean, tomato and potato. The right is gene maps of the same NAC TFs, showing the numbers of exon and intron, the absence or presence of upstream/downstream sequences and their relative length.
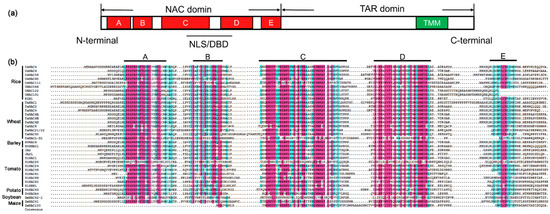
Figure 2. Diagram of the NAC proteins structure and sequence alignment of NAC domains. (a) Schematic representation showing a typical NAC protein with a highly conserved NAC domain at the N-terminal which is further divided into five conserved subdomains (A–E). The NAC domain contains nuclear localization signals (NLSs) and DNA-binding domain (DBD). The C-terminal region is a relatively divergent transcriptional activation region (TAR). In some cases, the C-terminal may have a transmembrane motif (TMM). (b) Sequence alignment of NAC domains for most of crops cited in the review. Subdomains A to E are shown by horizontal lines above the sequence. The red color represents amino acids with more than 75% common consensus sequences, and the cyan color represents amino acids with more than 50% common consensus sequences.
References
- Cohen, S.P.; Leach, J.E. Abiotic and biotic stresses induce a core transcriptome response in rice. Sci. Rep. 2019, 9, 6273, doi:10.1038/s41598-019-42731-8.
- Burdon, J.J.; Thrall, P.H.; Ericson, L. Plant Pathogens and Disease: Newly Emerging Diseases; Elsevier: Amsterdam, Netherlands, 2009.
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nat. Cell Biol. 2006, 444, 323–329, doi:10.1038/nature05286.
- Boller, T.; He, S.Y. Innate Immunity in Plants: An Arms Race between Pattern Recognition Receptors in Plants and Effectors in Microbial Pathogens. Science 2009, 324, 742–744, doi:10.1126/science.1171647.
- Zhang, J.; Zhou, J.-M. Plant Immunity Triggered by Microbial Molecular Signatures. Mol. Plant 2010, 3, 783–793, doi:10.1093/mp/ssq035.
- Dodds, P.N.; Rathjen, J.P. Plant immunity: Towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 2010, 11, 539–548, doi:10.1038/nrg2812.
- Miller, G.; Shulaev, V.; Mittler, R. Reactive oxygen signaling and abiotic stress. Physiol. Plant. 2008, 133, 481–489, doi:10.1111/j.1399-3054.2008.01090.x.
- Bari, R.; Jones, J.D.G. Role of plant hormones in plant defence responses. Plant Mol. Biol. 2009, 69, 473–488, doi:10.1007/s11103-008-9435-0.
- Munné-Bosch, S.; Queval, G.; Foyer, C.H. The Impact of Global Change Factors on Redox Signaling Underpinning Stress Tolerance. Plant Physiol. 2013, 161, 5–19, doi:10.1104/pp.112.205690.
- Martí, M.C.; Stancombe, M.A.; Webb, A.A.R. Cell- and Stimulus Type-Specific Intracellular Free Ca2+ Signals in Arabidopsis. Plant Physiol. 2013, 163, 625–634, doi:10.1104/pp.113.222901.
- Sakuraba, Y.; Kim, D.; Han, S.-H.; Kim, S.-H.; Piao, W.; Yanagisawa, S.; An, G.; Paek, N.-C. Multilayered Regulation of Membrane-Bound ONAC054 Is Essential for Abscisic Acid-Induced Leaf Senescence in Rice. Plant Cell 2020, 32, 630–649, doi:10.1105/tpc.19.00569.
- Mao, C.J.; He, J.M.; Liu, L.N.; Deng, Q.M.; Yao, X.F.; Liu, C.M.; Qiao, Y.L.; Li, P.; Ming, F. OsNAC2 integrates auxin and cyto-kinin pathways to modulate rice root development. Plant Biotechnol. J. 2020, 18, 429–442, doi:10.1111/pbi.13209.
- Jiang, D.; Zhou, L.; Chen, W.; Ye, N.; Xia, J.; Zhuang, C. Overexpression of a microRNA-targeted NAC transcription factor improves drought and salt tolerance in Rice via ABA-mediated pathways. Rice 2019, 12, 76.
- Ye, Y.F.; Wu, K.; Chen, J.F.; Liu, Q.; Wu, Y.J.; Liu, B.M.; Fu, X.D. OsSND2, a NAC family transcription factor, is involved in secondary cell wall biosynthesis through regulating MYBs expression in rice. Rice 2018, 11, 1–14, doi:10.1186/s12284-018-0228-z.
- Mao, H.; Li, S.; Wang, Z.; Cheng, X.; Li, F.; Mei, F.; Chen, N.; Kang, Z. Regulatory changes in TaSNAC8‐6A are associated with drought tolerance in wheat seedlings. Plant Biotechnol. J. 2020, 18, 1078–1092, doi:10.1111/pbi.13277.
- Ren, Z.; Zhang, D.; Cao, L.; Zhang, W.; Zheng, H.; Liu, Z.; Han, S.; Dong, Y.; Zhu, F.; Liu, H.; et al. Functions and regulatory framework of ZmNST3 in maize under lodging and drought stress. Plant Cell Environ. 2020, 43, 2272–2286, doi:10.1111/pce.13829.
- Li, S.; Wang, N.; Ji, D.D.; Zhang, W.X.; Wang, Y.; Yu, Y.C.; Zhao, S.Z.; Lyu, M.H.; You, J.J.; Zhang, Y.Y.; et al. A GmSIN1/GmNCED3s/GmRbohBs Feed-Forward Loop Acts as a Signal Amplifier That Regulates Root Growth in Soybean Exposed to Salt Stress. Plant Cell 2019, 31, 2107–2130.
- Ji, Q.; Xu, X.; Wang, K. Genetic transformation of major cereal crops. Int. J. Dev. Biol. 2013, 57, 495–508, doi:10.1387/ijdb.130244kw.
- Nuruzzaman, M.; Manimekalai, R.; Sharoni, A.M.; Satoh, K.; Kondoh, H.; Ooka, H.; Kikuchi, S. Genome-wide analysis of NAC transcription factor family in rice. Gene 2010, 465, 30–44, doi:10.1016/j.gene.2010.06.008.
- Aida, M.; Ishida, T.; Fukaki, H.; Fujisawa, H.; Tasaka, M. Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell 1997, 9, 841–857, doi:10.1105/tpc.9.6.841.
- Souer, E.; Van Houwelingen, A.; Kloos, D.; Mol, J.; Koes, R. The No Apical Meristem Gene of Petunia Is Required for Pattern Formation in Embryos and Flowers and Is Expressed at Meristem and Primordia Boundaries. Cell 1996, 85, 159–170, doi:10.1016/s0092-8674(00)81093-4.
- Lu, M.; Sun, Q.P.; Zhang, D.F.; Wang, T.Y.; Pan, J.B. Identification of 7 stress-related NAC transcription factor members in maize (Zea mays L.) and characterization of the expression pattern of these genes. Biochem. Biophys. Res. Commun. 2015, 462, 144–150, doi:10.1016/j.bbrc.2015.04.113.
- Lv, S.; Guo, H.; Zhang, M.; Wang, Q.; Zhang, H.; Ji, W. Large-Scale Cloning and Comparative Analysis of TaNAC Genes in Response to Stripe Rust and Powdery Mildew in Wheat (Triticum aestivum L.). Genes 2020, 11, 1073, doi:10.3390/genes11091073.
- Jamar, C.; Loffet, F.; Frettinger, P.; Ramsay, L.; Fauconnier, M.-L.; Du Jardin, P. NAM-1gene polymorphism and grain pro-tein content in Hordeum. J. Plant Physiol. 2010, 167, 497–501, doi:10.1016/j.jplph.2009.10.014.
- Le, D.T.; Nishiyama, R.; Watanabe, Y.; Mochida, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.-S.P. Genome-Wide Survey and Expression Analysis of the Plant-Specific NAC Transcription Factor Family in Soybean During Development and Dehydration Stress. DNA Res. 2011, 18, 263–276, doi:10.1093/dnares/dsr015.
- Jin, J.F.; Wang, Z.Q.; He, Q.Y.; Wang, J.Y.; Li, P.F.; Xu, J.M.; Zheng, S.J.; Fan, W.; Yang, J.L. Genome-wide identification and expression analysis of the NAC transcription factor family in tomato (Solanum lycopersicum) during aluminum stress. BMC Genom. 2020, 21, 1–14, doi:10.1186/s12864-020-6689-7.
- Mathew, I.E.; Agarwal, P. May the Fittest Protein Evolve: Favoring the Plant-Specific Origin and Expansion of NAC Tran-scription Factors. BioEssays 2018, 40, e1800018, doi:10.1002/bies.201800018.
- Marques, D.N.; Dos Reis, S.P.; De Souza, C.R.B. Plant NAC transcription factors responsive to abiotic stresses. Plant Gene 2017, 11, 170–179, doi:10.1016/j.plgene.2017.06.003.
- Yuan, X.; Wang, H.; Cai, J.; Li, D.; Song, F. NAC transcription factors in plant immunity. Phytopathol. Res. 2019, 1, 3, doi:10.1186/s42483-018-0008-0.
- Zhu, T.; Nevo, E.; Sun, D.; Peng, J. Phylogenetic Analyses Unravel the Evolutionary History of NAC Proteins in Plants. Evo-lution 2012, 66, 1833–1848, doi:10.1111/j.1558-5646.2011.01553.x.
- Pereira-Santana, A.; Alcaraz, L.D.; Castano, E.; Sánchez-Calderón, L.; Sanchez-Teyer, F.; Rodríguez-Zapata, L. Comparative Genomics of NAC Transcriptional Factors in Angiosperms: Implications for the Adaptation and Diversification of Flower-ing Plants. PLoS ONE 2015, 10, e0141866, doi:10.1371/journal.pone.0141866.
- Mohanta, T.K.; Yadav, D.; Khan, A.; Hashem, A.; Tabassum, B.; Khan, A.L.; Allah, E.F.; Al-Harrasi, A. Genomics, molecular and evolutionary perspective of NAC transcription factors. PLoS ONE 2020, 15, e0231425, doi:10.1371/journal.pone.0231425.
- Timme, R.E.; Bachvaroff, T.R.; Delwiche, C.F. Broad Phylogenomic Sampling and the Sister Lineage of Land Plants. PLoS ONE 2012, 7, e29696, doi:10.1371/journal.pone.0029696.
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10.
- Puranik, S.; Sahu, P.P.; Mandal, S.N.; B., V.S.; Parida, S.K.; Prasad, M. Comprehensive Genome-Wide Survey, Genomic Con-stitution and Expression Profiling of the NAC Transcription Factor Family in Foxtail Millet (Setaria italica L.). PLoS ONE 2013, 8, e64594, doi:10.1371/journal.pone.0064594.
- Jin, X.; Ren, J.; Nevo, E.; Yin, X.; Sun, D.; Peng, J. Divergent Evolutionary Patterns of NAC Transcription Factors Are Associ-ated with Diversification and Gene Duplications in Angiosperm. Front. Plant Sci. 2017, 8, 1156, doi:10.3389/fpls.2017.01156.
- Shang, H.; Li, W.; Zou, C.; Yuan, Y. Analyses of the NAC Transcription Factor Gene Family in Gossypium raimondii Ulbr.: Chromosomal Location, Structure, Phylogeny, and Expression Patterns. J. Integr. Plant Biol. 2013, 55, 663–676, doi:10.1111/jipb.12085.
- Hasson, A.; Plessis, A.; Blein, T.; Adroher, B.; Grigg, S.; Tsiantis, M.; Boudaoud, A.; Damerval, C.; Laufs, P. Evolution and Diverse Roles of the CUP-SHAPED COTYLEDON Genes in Arabidopsis Leaf Development. Plant Cell 2011, 23, 54–68, doi:10.1105/tpc.110.081448.
- Zhong, R.; Lee, C.; Ye, Z.H. Evolutionary conservation of the transcriptional network regulating secondary cell wall bio-synthesis. Trends Plant Sci. 2010, 15, 625–632, doi:10.1016/j.tplants.2010.08.007.
- Xu, B.; Ohtani, M.; Yamaguchi, M.; Toyooka, K.; Wakazaki, M.; Sato, M.; Kubo, M.; Nakano, Y.; Sano, R.; Hiwatashi, Y.; et al. Contribution of NAC Transcription Factors to Plant Adaptation to Land. Sci. 2014, 343, 1505–1508, doi:10.1126/science.1248417.
- Seo, P.J.; Kim, S.G.; Park, C.M., Membrane-bound transcription factors in plants. Trends Plant Sci. 2008, 13, 555–556.
- Liang, M.; Li, H.; Zhou, F.; Li, H.; Liu, J.; Hao, Y.; Wang, Y.; Zhao, H.; Han, S. Subcellular Distribution of NTL Transcription Factors in Arabidopsis thaliana. Traffic 2015, 16, 1062–1074, doi:10.1111/tra.12311.
- Kim, S.Y.; Kim, S.G.; Kim, Y.S.; Seo, P.J.; Bae, M.; Yoon, H.K.; Park, C.M. Exploring membrane-associated NAC transcription factors in Arabidopsis : Implications for membrane biology in genome regulation. Nucleic Acids Res. 2006, 35, 203–213, doi:10.1093/nar/gkl1068.
- Kikuchi, K.; Ueguchi-Tanaka, M.; Yoshida, K.T.; Nagato, Y.; Matsusoka, M.; Hirano, H.-Y. Molecular analysis of the NAC gene family in rice. Mol. Genet. Genom. 2000, 262, 1047–1051, doi:10.1007/pl00008647.
- Jensen, M.K.; Kjaersgaard, T.; Nielsen, M.M.; Galberg, P.; Petersen, K.; O’Shea, C.; Skriver, K. The Arabidopsis thaliana NAC transcription factor family: Structure–function relationships and determinants of ANAC019 stress signalling. Biochem. J. 2010, 426, 183–196, doi:10.1042/bj20091234.
- Puranik, S.; Sahu, P.P.; Srivastava, P.S.; Prasad, M. NAC proteins: Regulation and role in stress tolerance. Trends Plant Sci. 2012, 17, 369–381, doi:10.1016/j.tplants.2012.02.004.
- Olsen, A.N.; Ernst, H.A.; Leggio, L.L.; Skriver, K. NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci. 2005, 10, 79–87, doi:10.1016/j.tplants.2004.12.010.
- Ooka, H.; Satoh, K.; Doi, K.; Nagata, T.; Otomo, Y.; Murakami, K.; Matsubara, K.; Osato, N.; Kawai, J.; Carninci, P.; et al. Comprehensive Analysis of NAC Family Genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 2003, 10, 239–247, doi:10.1093/dnares/10.6.239.
- Ernst, H.A.; Olsen, A.N.; Skriver, K.; Larsen, S.; Lo Leggio, L. Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors. EMBO Rep. 2004, 5, 297–303, doi:10.1038/sj.embor.7400093.
- Chen, Q.; Wang, Q.; Xiong, L.; Lou, Z. A structural view of the conserved domain of rice stress-responsive NAC1. Protein Cell 2011, 2, 55–63, doi:10.1007/s13238-011-1010-9.
- Welner, D.H.; Lindemose, S.; Grossmann, J.G.; Møllegaard, N.E.; Olsen, A.N.; Helgstrand, C.; Skriver, K.; Lo Leggio, L. DNA binding by the plant-specific NAC transcription factors in crystal and solution: A firm link to WRKY and GCM transcrip-tion factors. Biochem. J. 2012, 444, 395–404, doi:10.1042/BJ20111742.
- Shen, H.; Yin, Y.; Chen, F.; Xu, Y.; A Dixon, R. A Bioinformatic Analysis of NAC Genes for Plant Cell Wall Development in Relation to Lignocellulosic Bioenergy Production. BioEnergy Res. 2009, 2, 217–232, doi:10.1007/s12155-009-9047-9.
- Delessert, C.; Kazan, K.; Wilson, I.W.; Van Der Straeten, D.; Manners, J.; Dennis, E.S.; Dolferus, R. The transcription factor ATAF2 represses the expression of pathogenesis-related genes in Arabidopsis. Plant J. 2005, 43, 745–757, doi:10.1111/j.1365-313x.2005.02488.x.
- Wang, X.E.; Basnayake, B.M.V.S.; Zhang, H.J.; Li, G.J.; Li, W.; Virk, N.; Mengiste, T.; Song, F.M. The Arabidopsis ATAF1, a NAC Transcription Factor, Is a Negative Regulator of Defense Responses Against Necrotrophic Fungal and Bacterial Path-ogens. Mol. Plant-Microbe Interact. 2009, 22, 1227–1238, doi:10.1094/mpmi-22-10-1227.
- Wu, Y.R.; Deng, Z.Y.; Lai, J.B.; Zhang, Y.Y.; Yang, C.P.; Yin, B.J.; Zhao, Q.Z.; Zhang, L.; Li, Y.; Yang, C.W.; et al. Dual function of Arabidopsis ATAF1 in abiotic and biotic stress responses. Cell Res. 2009, 19, 1279–1290, doi:10.1038/cr.2009.108.
- Huang, W.Z.; Miao, M.; Kud, J.N.; Niu, X.L.; Ouyang, B.; Zhang, J.H.; Ye, Z.B.; Kuhl, J.C.; Liu, Y.S.; Xiao, F.M. SlNAC1, a stress-related transcription factor, is fine-tuned on both the transcriptional and the post-translational level. New Phytol. 2012, 197, 1214–1224, doi:10.1111/nph.12096.
- Zhang, Z.X.; Zhang, Y.P.; Zhao, H.; Huang, F.L.; Zhang, Z.; Lin, W.X. The important functionality of 14-3-3 isoforms in rice roots revealed by affinity chromatography. J. Proteom. 2017, 158, 20–30, doi:10.1016/j.jprot.2017.02.008.
- Nakashima, K.; Tran, L.S.; Van Nguyen, D.; Fujita, M.; Maruyama, K.; Todaka, D.; Ito, Y.; Hayashi, N.; Shinozaki, K.; Yama-guchi-Shinozaki, K. Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J. 2007, 51, 617–630, doi:10.1111/j.1365-313x.2007.03168.x.
- Yokotani, N.; Tsuchida-Mayama, T.; Ichikawa, H.; Mitsuda, N.; Ohme-Takagi, M.; Kaku, H.; Minami, E.; Nishizawa, Y. Os-NAC111, a Blast Disease–Responsive Transcription Factor in Rice, Positively Regulates the Expression of Defense-Related Genes. Mol. Plant-Microbe Interact. 2014, 27, 1027–1034, doi:10.1094/mpmi-03-14-0065-r.
- Park, S.R.; Kim, H.S.; Lee, K.S.; Hwang, D.J.; Kim, S.T. Overexpression of rice NAC transcription factor OsNAC58 on in-creased resistance to bacterial leaf blight. J. Plant Biotechnol. 2017, 44, 149–155, doi:10.5010/jpb.2017.44.2.149.
- Sun, L.; Zhang, H.; Li, D.; Huang, L.; Hong, Y.; Ding, X.S.; Nelson, R.S.; Zhou, X.; Song, F. Functions of rice NAC transcrip-tional factors, ONAC122 and ONAC131, in defense responses against Magnaporthe grisea. Plant Mol. Biol. 2013, 81, 41–56, doi:10.1007/s11103-012-9981-3.
- Liu, Q.; Yan, S.; Huang, W.; Yang, J.; Dong, J.; Zhang, S.; Zhao, J.; Yang, T.; Mao, X.; Zhu, X.; et al. NAC transcription factor ONAC066 positively regulates disease resistance by suppressing the ABA signaling pathway in rice. Plant Mol. Biol. 2018, 98, 289–302, doi:10.1007/s11103-018-0768-z.
- Kaneda, T.; Taga, Y.; Takai, R.; Iwano, M.; Matsui, H.; Takayama, S.; Isogai, A.; Che, F.-S. The transcription factor OsNAC4 is a key positive regulator of plant hypersensitive cell death. EMBO J. 2009, 28, 926–936, doi:10.1038/emboj.2009.39.
- Wang, Z.; Xia, Y.; Lin, S.; Wang, Y.; Guo, B.; Song, X.; Ding, S.; Zheng, L.; Feng, R.; Chen, S.; et al. Osa-miR164a targets Os-NAC60 and negatively regulates rice immunity against the blast fungus Magnaporthe oryzae. Plant J. 2018, 95, 584–597.
- Yoshii, M.; Shimizu, T.; Yamazaki, M.; Higashi, T.; Miyao, A.; Hirochika, H.; Omura, T. Disruption of a novel gene for a NAC-domain protein in rice confers resistance toRice dwarf virus. Plant J. 2009, 57, 615–625, doi:10.1111/j.1365-313x.2008.03712.x.
- Bell, G.D.H. The History of Wheat Cultivation; Springer: Amsterdam, The Netherlands, 1987.
- Kinnunen, P.; Guillaume, J.H.A.; Taka, M.; D’Odorico, P.; Siebert, S.; Puma, M.J.; Jalava, M.; Kummu, M. Local food crop production can fulfil demand for less than one-third of the population. Nat. Food 2020, 1, 229–237, doi:10.1038/s43016-020-0060-7.
- Qian, C.; Cui, C.; Wang, X.; Zhou, C.; Hu, P.; Li, M.; Li, R.; Xiao, J.; Wang, X.; Chen, P.; Xing, L.; Cao, A. Molecular character-isation of the broad-spectrum resistance to powdery mildew conferred by the Stpk-V gene from the wild species Haynaldia villosa. Plant Biol. 2017, 19, 875–885.
- Zhou, W.H.; Qian, C.; Li, R.C.; Zhou, S.; Zhang, R.Q.; Xiao, J.; Wang, X.E.; Zhang, S.Z.; Xing, L.P.; Cao, A.Z. TaNAC6s are involved in the basal and broad-spectrum resistance to powdery mildew in wheat. Plant Sci. 2018, 277, 218–228, doi:10.1016/j.plantsci.2018.09.014.
- Perochon, A.; Kahla, A.; Vranić, M.; Jia, J.G.; Malla, K.B.; Craze, M.; Wallington, E.J.; Doohan, F.M. A wheat NAC interacts with an orphan protein and enhances resistance to Fusarium head blight disease. Plant Biotechnol. J. 2019, 17, 1892–1904, doi:10.1111/pbi.13105.
- Perochon, A.; Jia, J.G.; Kahla, A.; Arunachalam, C.; Scofield, S.R.; Bowden, S.; Wallington, E.J.; Doohan, F.M. TaFROG en-codes a Pooideae orphan protein that interacts with SnRK1 and enhances resistance to the mycotoxigenic fungus Fusarium graminearum. Plant Physiol. 2015, 169, 2895–2906, doi:10.1104/pp.15.01056.
- Xia, N.; Zhang, G.; Sun, Y.F.; Zhu, L.; Xu, L.S.; Chen, X.M.; Liu, B.; Yu, Y.-T.; Wang, X.-J.; Huang, L.L.; et al. TaNAC8, a novel NAC transcription factor gene in wheat, responds to stripe rust pathogen infection and abiotic stresses. Physiol. Mol. Plant Pathol. 2010, 74, 394–402, doi:10.1016/j.pmpp.2010.06.005.
- Xia, N.; Zhang, G.; Liu, X.-Y.; Deng, L.; Cai, G.-L.; Zhang, Y.; Wang, X.-J.; Zhao, J.; Huang, L.-L.; Kang, Z.S. Characterization of a novel wheat NAC transcription factor gene involved in defense response against stripe rust pathogen infection and abiotic stresses. Mol. Biol. Rep. 2010, 37, 3703–3712, doi:10.1007/s11033-010-0023-4.
- Feng, H.; Duan, X.; Zhang, Q.; Li, X.; Wang, B.; Huang, L.; Wang, X.; Kang, Z. The target gene of tae-miR164, a novel NAC transcription factor from the NAM subfamily, negatively regulates resistance of wheat to stripe rust. Mol. Plant Pathol. 2014, 15, 284–296, doi:10.1111/mpp.12089.
- Wang, F.T.; Lin, R.M.; Feng, J.; Chen, W.Q.; Qiu, D.W.; Xu, S.C. TaNAC1 acts as a negative regulator of stripe rust resistance in wheat, enhances susceptibility to Pseudomonas syringae, and promotes lateral root development in transgenic Ara-bidopsis thaliana. Front. Plant Sci. 2015, 6, 108, doi:10.3389/fpls.2015.00108.
- Zhang, X.M.; Zhang, Q.; Pei, C.L.; Li, X.; Huang, X.L.; Chang, C.-Y.; Wang, X.J.; Huang, L.L.; Kang, Z.S. TaNAC2 is a negative regulator in the wheat-stripe rust fungus interaction at the early stage. Physiol. Mol. Plant Pathol. 2018, 102, 144–153, doi:10.1016/j.pmpp.2018.02.002.
- Wang, B.; Wei, J.; Song, N.; Wang, N.; Zhao, J.; Kang, Z. A novel wheat NAC transcription factor, TaNAC30, negatively reg-ulates resistance of wheat to stripe rust. J. Integr. Plant Biol. 2018, 60, 432–443.
- Newton, A.C.; Flavell, A.J.; George, T.S.; Leat, P.; Mullholland, B.; Ramsay, L.; Revoredo-Giha, C.; Russell, J.; Steffenson, B.J.; Swanston, J.S.; et al. Crops that feed the world Barley: A resilient crop? Strengths and weaknesses in the context of food se-curity. Food Secur. 2011, 3, 141–178, doi:10.1007/s12571-011-0126-3.
- Walters, D.R.; Avrova, A.; Bingham, I.J.; Burnett, F.J.; Fountaine, J.; Havis, N.D.; Hoad, S.P.; Hughes, G.; Looseley, M.; Oxley, S.J.P.; et al. Control of foliar diseases in barley: Towards an integrated approach. Eur. J. Plant Pathol. 2012, 133, 33–73, doi:10.1007/s10658-012-9948-x.
- Christiansen, M.W.; Holm, P.B.; Gregersen, P.L. Characterization of barley (Hordeum vulgare L.) NAC transcription factors suggests conserved functions compared to both monocots and dicots. BMC Res. Notes 2011, 4, 302, doi:10.1186/1756-0500-4-302.
- Chen, Y.J.; Perera, V.; Christiansen, M.W.; Holme, I.B.; Gregersen, P.L.; Grant, M.R.; Collinge, D.B.; Lyngkjær, M.F. The bar-ley HvNAC6 transcription factor affects ABA accumulation and promotes basal resistance against powdery mildew. Plant Mol. Biol. 2013, 83, 577–590, doi:10.1007/s11103-013-0109-1.
- Jensen, M.K.; Rung, J.H.; Gregersen, P.L.; Gjetting, T.; Fuglsang, A.T.; Hansen, M.; Joehnk, N.; Lyngkjaer, M.F.; Collinge, D.B. The HvNAC6 transcription factor: A positive regulator of penetration resistance in barley and Arabidopsis. Plant Mol. Biol. 2007, 65, 137–150, doi:10.1007/s11103-007-9204-5.
- McGrann, G.R.; Steed, A.; Burt, C.; Goddard, R.; Lachaux, C.; Bansal, A.; Corbitt, M.; Gorniak, K.; Nicholson, P.; Brown, J.K. Contribution of the drought tolerance-relatedStress-responsive NAC1transcription factor to resistance of barley to Ramu-laria leaf spot. Mol. Plant Pathol. 2015, 16, 201–209, doi:10.1111/mpp.12173.
- Walters, D.R.; Havis, N.D.; Oxley, S.J. Ramularia collo-cygni: The biology of an emerging pathogen of barley. FEMS Micro-biol. Lett. 2008, 279, 1–7, doi:10.1111/j.1574-6968.2007.00986.x.
- Zsogon, A.; Cermak, T.; Naves, E.R.; Notini, M.M.; Edel, K.H.; Weinl, S.; Freschi, L.; Voytas, D.F.; Kudla, J.; Peres, L.E.P. De novo domestication of wild tomato using genome editing. Nat. Biotechnol. 2018, 36, 1211–1216.
- Li, T.D.; Yang, X.P.; Yu, Y.; Si, X.M.; Zhai, X.W.; Zhang, H.W.; Dong, W.X.; Gao, C.X.; Xu, C. Domestication of wild tomato is accelerated by genome editing. Nat. Biotechnol. 2018, 36, 1160–1163.
- Zhu, G.T.; Wang, S.C.; Huang, Z.J.; Zhang, S.B.; Liao, Q.G.; Zhang, C.Z.; Lin, T.; Qin, M.; Peng, M.; Yang, C.K.; et al. Rewiring of the Fruit Metabolome in Tomato Breeding. Cell 2018, 172, 249–261.
- Baysal-Gurel, F.; Li, R.; Ling, K.S.; Miller, S.A. First Report of Tomato chlorotic spot virus Infecting Tomatoes in Ohio. Plant Dis. 2015, 99, 163, doi:10.1094/pdis-06-14-0639-pdn.
- Balmant, K.M.; Parker, J.; Yoo, M.J.; Zhu, N.; Dufresne, C.; Chen, S.X. Redox proteomics of tomato in response to Pseudo-monas syringae infection. Hortic. Res. 2015, 2, 15043, doi:10.1038/hortres.2015.43.
- Bhattarai, K.; Louws, F.J.; Williamson, J.D.; Panthee, D.R. Differential response of tomato genotypes to Xanthomo-nas-specific pathogen-associated molecular patterns and correlation with bacterial spot (Xanthomonas perforans) resistance. Hortic. Res. 2016, 3, 16035, doi:10.1038/hortres.2016.35.
- Han, Q.; Zhang, J.; Li, H.; Luo, Z.; Ziaf, K.; Ouyang, B.; Wang, T.; Ye, Z. Identification and expression pattern of one stress-responsive NAC gene from Solanum lycopersicum. Mol. Biol. Rep. 2011, 39, 1713–1720, doi:10.1007/s11033-011-0911-2.
- Ma, N.N.; Zuo, Y.Q.; Liang, X.Q.; Yin, B.; Wang, G.D.; Meng, Q.W. The multiple stress-responsive transcription factor SlNAC1 improves the chilling tolerance of tomato. Physiol. Plant. 2013, 149, 474–486.
- Zhu, M.; Chen, G.; Zhou, S.; Tu, Y.; Wang, Y.; Dong, T.; Hu, Z. A New Tomato NAC (NAM/ATAF1/2/CUC2) Transcription Factor, SlNAC4, Functions as a Positive Regulator of Fruit Ripening and Carotenoid Accumulation. Plant Cell Physiol. 2013, 55, 119–135, doi:10.1093/pcp/pct162.
- Huang, Y.; Li, T.; Xu, Z.-S.; Wang, F.; Xiong, A.-S. Six NAC transcription factors involved in response to TYLCV infection in resistant and susceptible tomato cultivars. Plant Physiol. Biochem. 2017, 120, 61–74, doi:10.1016/j.plaphy.2017.09.020.
- Cottilli, P.; Belda-Palazón, B.; Adkar-Purushothama, C.R.; Perreault, J.-P.; Schleiff, E.; Rodrigo, I.; Ferrando, A.; Lison, P. Citrus exocortis viroid causes ribosomal stress in tomato plants. Nucleic Acids Res. 2019, 47, 8649–8661, doi:10.1093/nar/gkz679.
- Selth, L.A.; Dogra, S.C.; Rasheed, M.S.; Healy, H.; Randles, J.W.; Rezaian, M.A. A NAC Domain Protein Interacts with To-mato leaf curl virus Replication Accessory Protein and Enhances Viral Replication. Plant Cell 2005, 17, 311–325, doi:10.1105/tpc.104.027235.
- Miao, M.; Niu, X.L.; Kud, J.; Du, X.R.; Avila, J.; Devarenne, T.P.; Kuhl, J.C.; Liu, Y.S.; Xiao, F.M. The ubiquitin ligase SEVEN IN ABSENTIA (SINA) ubiquitinates a defense-related NAC transcription factor and is involved in defense signaling. New Phytol. 2016, 211, 138–148.
- Liu, B.; Ouyang, Z.; Zhang, Y.; Li, X.; Hong, Y.; Huang, L.; Liu, S.; Zhang, H.; Li, D.; Song, F. Tomato NAC Transcription Fac-tor SlSRN1 Positively Regulates Defense Response against Biotic Stress but Negatively Regulates Abiotic Stress Response. PLoS ONE 2014, 9, e102067, doi:10.1371/journal.pone.0102067.
- Arnaud, D.; Hwang, I. A sophisticated network of signaling pathways regulates stomatal defenses to bacterial pathogens. Mol. Plant 2014, 8, 566–581, doi:10.1093/mp/ssu129.
- Du, M.; Zhai, Q.; Deng, L.; Li, S.; Li, H.; Yan, L.; Huang, Z.; Wang, B.; Jiang, H.; Huang, T.; et al. Closely Related NAC Tran-scription Factors of Tomato Differentially Regulate Stomatal Closure and Reopening during Pathogen Attack. Plant Cell 2014, 26, 3167–3184, doi:10.1105/tpc.114.128272.
- Chang, Y.; Yu, R.; Feng, J.; Chen, H.; Eri, H.; Gao, G. NAC transcription factor involves in regulating bacterial wilt re-sistance in potato. Funct. Plant Biol. 2020, 47, 925, doi:10.1071/fp19331.
- Yogendra, K.N.; Sarkar, K.; Kage, U.; Kushalappa, A.C. Potato NAC43 and MYB8 Mediated Transcriptional Regulation of Secondary Cell Wall Biosynthesis to Contain Phytophthora infestans Infection. Plant Mol. Biol. Rep. 2017, 35, 519–533, doi:10.1007/s11105-017-1043-1.
- Schnable, J.C. Genome Evolution in Maize: From Genomes Back to Genes. Annu. Rev. Plant Biol. 2015, 66, 329–343, doi:10.1146/annurev-arplant-043014-115604.
- Li, W.; Deng, Y.; Ning, Y.; He, Z.; Wang, G.L. Exploiting Broad-Spectrum Disease Resistance in Crops: From Molecular Dis-section to Breeding. Annu. Rev. Plant Biol. 2020, 71, 575–603, doi:10.1146/annurev-arplant-010720-022215.
- Peng, X.; Zhao, Y.; Li, X.; Wu, M.; Chai, W.; Sheng, L.; Wang, Y.; Dong, Q.; Jiang, H.; Cheng, B. Genomewide identification, classification and analysis of NAC type gene family in maize. J. Genet. 2015, 94, 377–390, doi:10.1007/s12041-015-0526-9.
- Voitsik, A.M.; Münch, S.; Deising, H.B.; Voll, L.M. Two recently duplicated maize NAC transcription factor paralogs are induced in response to Colletotrichum graminicola infection. BMC Plant Biol. 2013, 13, 1–16, doi:10.1186/1471-2229-13-85.
- Shoemaker, R.C.; Polzin, K.; Labate, J.; Specht, J.; Brummer, E.C.; Olson, T.; Young, N.; Concibido, V.; Wilcox, J.; Tamulonis, J.P.; et al. Genome Duplication in Soybean (Glycine Subgenus Soja). Genetics 1996, 144, 329–338.
- Meng, Q.; Zhang, C.; Gai, J.; Yu, D. Molecular cloning, sequence characterization and tissue-specific expression of six NAC-like genes in soybean (Glycine max (L.) Merr.). J. Plant Physiol. 2007, 164, 1002–1012, doi:10.1016/j.jplph.2006.05.019.
- Pinheiro, G.L.; Marques, C.S.; Costa, M.D.; Reis, P.A.; Alves, M.S.; Carvalho, C.M.; Fietto, L.G.; Fontes, E.P.B. Complete in-ventory of soybean NAC transcription factors: Sequence conservation and expression analysis uncover their distinct roles in stress response. Gene 2009, 444, 10–23, doi:10.1016/j.gene.2009.05.012.
- Mendes, G.C.; Reis, P.A.B.; Calil, I.P.; Carvalho, H.H.; Aragão, F.J.L.; Fontes, E.P.B. GmNAC30 and GmNAC81 integrate the endoplasmic reticulum stress- and osmotic stress-induced cell death responses through a vacuolar processing enzyme. Proc. Natl. Acad. Sci. USA 2013, 110, 19627–19632, doi:10.1073/pnas.1311729110.
- Pimenta, M.R.; Silva, P.A.; Mendes, G.C.; Alves, J.R.; Caetano, H.D.N.; Machado, J.P.B.; Brustolini, O.J.B.; Carpinetti, P.A.; Melo, B.P.; Silva, J.C.F.; et al. The Stress-Induced Soybean NAC Transcription Factor GmNAC81 Plays a Positive Role in Developmentally Programmed Leaf Senescence. Plant Cell Physiol. 2016, 57, 1098–1114.
- Hoang, X.L.T.; Nguyen, N.C.; Nguyen, Y.N.H.; Watanabe, Y.; Tran, L.S.P.; Thao, N.P. The Soybean GmNAC019 Transcription Factor Mediates Drought Tolerance in Arabidopsis in an Abscisic Acid-Dependent Manner. Int. J. Mol. Sci. 2019, 21, 286, doi:10.3390/ijms21010286.
- Yang, C.; Huang, Y.; Lv, W.; Zhang, Y.; Bhat, J.A.; Kong, J.; Xing, H.; Zhao, J.; Zhao, T. GmNAC8 acts as a positive regulator in soybean drought stress. Plant Sci. 2020, 293, 110442, doi:10.1016/j.plantsci.2020.110442.
- Jahan, M.A.; Harris, B.; Lowery, M.; Coburn, K.; Infante, A.M.; Percifield, R.J.; Ammer, A.G.; Kovinich, N. The NAC family transcription factor GmNAC42–1 regulates biosynthesis of the anticancer and neuroprotective glyceollins in soybean. BMC Genom. 2019, 20, 149, doi:10.1186/s12864-019-5524-5.
- Jahan, M.A.; Kovinich, N. Acidity stress for the systemic elicitation of glyceollin phytoalexins in soybean plants. Plant Signal. Behav. 2019, 14, 1604018, doi:10.1080/15592324.2019.1604018.
- Jahan, M.A.; Harris, B.; Lowery, M.; Infante, A.M.; Percifield, R.J.; Kovinich, N. Glyceollin Transcription Factor GmMYB29A2 Regulates Soybean Resistance to Phytophthora sojae. Plant Physiol. 2020, 183, 530–546, doi:10.1104/pp.19.01293.
- Meisrimler, C.N.; Pelgrom, A.J.E.; Oud, B.; Out, S.; Van den Ackerveken, G. Multiple downy mildew effectors target the stress-related NAC transcription factor LsNAC069 in lettuce. Plant J. 2019, 99, 1098–1115.




