
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | JYAWEI CHENG | + 1530 word(s) | 1530 | 2020-04-22 10:40:33 | | | |
| 2 | Rita Xu | Meta information modification | 1530 | 2020-04-27 09:40:42 | | | | |
| 3 | Rita Xu | -19 word(s) | 1511 | 2020-10-29 08:59:26 | | |
Video Upload Options
In the absence of proper immunity, such as in the case of acquired immune deficiency syndrome (AIDS) patients, Candida albicans, the most common human fungal pathogen, may cause mucosal and even life-threatening systemic infections. P-113 (AKRHHGYKRKFH), an antimicrobial peptide (AMP) derived from the human salivary protein histatin 5, shows good safety and efficacy profiles in gingivitis and human immunodeficiency virus (HIV) patients with oral candidiasis. However, little is known about how P-113 interacts with Candida albicans or its degradation by Candida-secreted proteases that contribute to the fungi’s resistance. Here, we use solution nuclear magnetic resonance (NMR) methods to elucidate the molecular mechanism of interactions between P-113 and living Candida albicans cells. Furthermore, we found that proteolytic cleavage of the C-terminus prevents the entry of P-113 into cells and that increasing the hydrophobicity of the peptide can significantly increase its antifungal activity.
1. NMR Time Course Study of P-113 Degradation by C. albicans
To determine the sequence of cleavage events of P-113, the time-dependent chemical shifts of P-113 incubated with C. albicans were recorded by NMR spectroscopy. Confocal and electron scanning micrographs were analyzed at the same time points to determine P-113 translocation and effects on cell morphology (Figure 1). To help identify the chemical shifts of each detected amino acid, the selective 15N-unlabeled peptides (Lys, Arg, and His residues) were used. New Lys8, Arg9, Lys10, and His12 cross-peaks were observed after 1 h, indicating that the first cleavage site was between the Lys10 and Phe11 residues. Therefore, hydrolysis of the Lys10-Phe11 peptide bond occurred first, releasing the C-terminal fragment. Furthermore, new cross-peaks were observed for His5, Gly6, Tyr7, Lys8, and Arg9 after 2–3 h of incubation. These results revealed that the farther cleavage site was between residues Tyr7 and Lys8. The original peaks from His5 to Phe11 with a gradual decay after 4 h of incubation indicated that most of the P-113 peptides were hydrolyzed by this time period. Furthermore, the increasing intensities of the new Arg9 cross-peak indicated that the cleavage site was located between Lys8 and Arg9 residues (Figure 1). Chemical shift perturbations were observed at 5 h of incubation (bottom panel of Figure 1). After 5 h of incubation with C. albicans, no further perturbation of chemical shifts was observed, which suggests a near-complete degradation of P-113. Although P-113 degradation started within an hour of incubation, peptides had already translocated into cells, as shown in confocal experiments (Figure 1). The effect of the peptides on cell morphology was also examined by scanning electron microscopy (SEM). The observed cell surfaces were smooth, which indicated that peptides penetrated cell membranes without causing disruption.
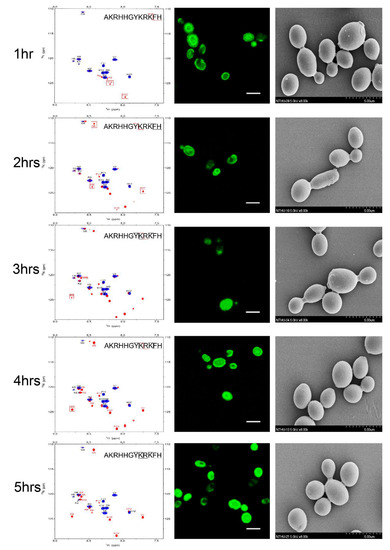
Figure 1. Time-course analysis of 0.25 mM P-113 cleavage following interaction with 107 CFU/mL of C. albicans by HSQC spectroscopy (left panels), confocal laser microscopy (middle panels), and scanning electron microscopy (right panels). The bar corresponds to 5 μm. An overlay of 1H-15N HSQC spectra for 0.25 mM of P-113 in SD broth only (blue) and broth with C. albicans at 301 K at different time points (red) is shown. Perturbations of chemical shifts were observed over 5 h of incubation with C. albicans cells. Cell morphology did not change over the 5 h incubation period.
2. Functional Role of N- and C-Terminal Amino Acids in the Antifungal Activity of P-113
Our results suggested that the lysine residues in the P-113 peptide sequence are the targets of cleavage by C. albicans. Interestingly, Lys2 is not hydrolyzed by the enzymes, which indicates that the N-terminal lysine has another function. To evaluate whether the N-terminal lysine residue is crucial for the activity of P-113 peptides, we removed the Ala–Lys residues at the N-terminal end of P-113 (N-terminal truncated P-113) and compared it with a P-113 peptide lacking Phe-His residues at the C-terminal end (C-terminal truncated P-113). All peptides were synthesized with fluorescein fluorescent (FITC) tagged at the N-terminal end and amidation at the C-terminal end. The antifungal activities of intact and truncated peptides were examined by two assays: a broth microdilution assay in which the minimum inhibitory concentration (MIC) values were determined and a time killing assay at 1× MIC. The MICs of the truncated P-113s were 8-16-fold higher than that of the full peptide (Table 1).
Table 1. Minimum inhibitory concentration (MIC) values of P-113 and its derivatives against C. albicans American Type Culture Collection (ATCC) 10231 (µg/mL). * The MIC of P-113 without the FITC label is the same as the MIC of FITC-P-113.
| Peptide | Sequence | Molecular Weight (Da) | MH Broth | LYM | ||
|---|---|---|---|---|---|---|
| Control | 50 mM NaCl | 100 mM NaCl | ||||
| P-113 * | FITC-AKRHHGYKRKFH-NH2 | 2066.36 | >50 | 3.13 | 25 | 50 |
| N-ter truncated P-113 | FITC-RHHGYKRKFH-NH2 | 1867.14 | >50 | 25 | >50 | >50 |
| C-ter truncated P-113 | FITC-AKRHHGYKRK-NH2 | 1781.49 | >50 | 50 | >50 | >50 |
| Bip-P-113 | FITC-AKR(Bip)(Bip)GYKRKF(Bip)-NH2 | 2324.62 | 50 | 6.25 | 6.25 | 12.5 |
| Dip-P-113 | FITC-AKR(Dip)(Dip)GYKRKF(Dip)-NH2 | 2324.62 | >50 | 12.5 | 12.5 | 25 |
Similarly, while treatment of C. albicans for 120 min with P-113 resulted in 87% cell killing, cell killing by N-terminal and C-terminal truncated peptides was 38% and 25%, respectively (Figure 2). Interestingly, C-terminal truncated P-113, which lacks the first fragment cleaved by C. albicans, killed the cells at much slower rates than N-terminal truncated P-113 (Figure 2a). These results, along with the MIC data, offer several insights into the different functional roles of N- and C-terminal amino acids in P-113 peptides.
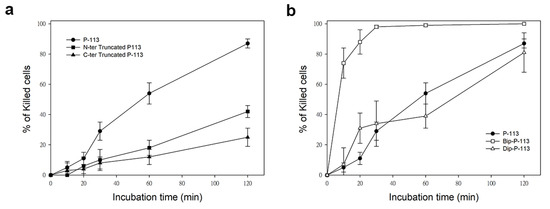
Figure 2. Time killing activity of FITC-labeled P-113 peptide and its FITC-labeled derivatives against Candida albicans. Cells were treated with 1× MIC of (a) P-113 (black circle), FITC-labeled N-terminal truncated P-113 (black square), FITC-labeled C-terminal truncated P-113 (black triangle), and (b) treated with FITC-labeled Bip-P-113 (white square), and FITC-labeled Dip-P-113 (white triangle). Killed cells (%) = (cell number in peptide-free control − cell number in sample)/(cell number in peptide-free control) × 100. Error bars represent the standard errors of the mean.
3. Substituting His 4, 5, and 12 of P-113 for Non-Natural Bulky Amino Acids Altered the Anti-Candida Mechanism
Previously, we found that the replacement of the aromatic residues with bulky aromatic amino acids may generate a potent peptide with improved antimicrobial activity and salt resistance [1]. We synthesized Bip-P-113 and Dip-P-113 by replacing the P-113 histidine residues (His4, His5, and His12) with β-(4,4β-biphenyl)alanine (Bip) and β-diphenylalanine (Dip), respectively. The peptides were conjugated with FITC, and the anti-Candida activities of P-113, Bip-P-113, and Dip-P-113 were determined in LYM broth with different salt concentrations (Table 1). Bip-P-113 and Dip-P-113 retained their anti-Candida activities under high-salt conditions, while the candidacidal activity of P-113 was reduced by the addition of 100 mM NaCl to the LYM medium. In addition to improved salt resistance, Bip-P-113 exhibited higher killing activity than the other two peptides under normal salt conditions, causing a 99% reduction in cell viability in 30 min, while P-113 and Dip-P-113 exhibited a 87% and 81% reduction after 120 min of incubation, respectively (Figure 2b). Interestingly, Dip-P-113 showed similar kinetics to P-113, which was translocated into the cells and then exhibited killing activity. Membrane disruptions and intracellular translocations of antimicrobial peptides are the two major mechanisms used by candidacidal targets. Next, we studied whether the antimicrobial activity of the bulky amino-acid replaced peptides involved Candida cell membrane disruption or translocation into the cytoplasm. Confocal microscopy imaging showed that, while P-113 accumulated in the cytosol after 5 min of incubation with Candida cells, Bip-P-113 and Dip-P-113 accumulated on the surface of cells and did not translocate into the cytoplasm (Figure 3a), suggesting that these peptides may exert their effect by direct membrane disruption. Therefore, we evaluated peptide-induced cellular membrane damage by SEM. Untreated C. albicans exhibited rough bright and smooth surfaces, whereas the membrane surfaces of C. albicans cells exposed to Bip-P-113 or Dip-P-113 for 5 min became rippled and showed deep pore formation (Figure 3b). These results demonstrate that the antifungal activities of Bip-P-113 and Dip-P-113 involve cell membrane disruption.
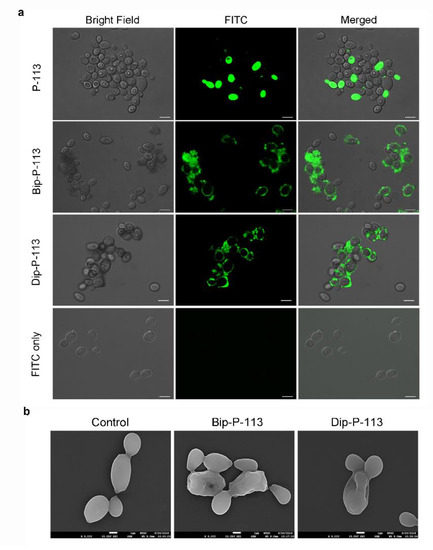
Figure 3. (a) Localizations of FITC peptides in in Candida albicans. Confocal microscopy was used to visualize 107 CFU/mL of C. albicans were incubated at 28 °C for 5 min with 50 μM of FITC-conjugated peptides. The left panels show a bright field, the middle panels show FITC images, and the right panels show merged images. The bar corresponds to 5 μm. (b) Scanning electron microscopic micrographs of C. albicans only as a control and treated with Bip-P-113 and Dip-P-113 peptides (Bip = β-(4,4β-biphenyl)alanine; Dip = β-diphenylalanine). Each figure is magnified ×8000.
4. Model of the proposed interactions of P-113 and Bip-P-113 with Candida albicans
We found that Phe-His residues at the C-terminus of P-113 play a pivotal role in peptide translocation, while the N-terminal Ala-Lys residues are key for anticandidal activity. The resistance strategy of C. albicans relies on the proteolytic defensive enzymes that specifically cleaved Phe–His amino-acid sequences, preventing peptides from importing into the cytosol. These findings provide further in-depth information for the design of effective anti-Candida agents. Furthermore, elongating the length of the hydrophobic alkyl tails at the position of histidine alters the C. albicans killing mechanism of P-113. With its high anti-Candida activity, Bip-P-113 deserves further attention in the development of antifungal therapeutics in the future.
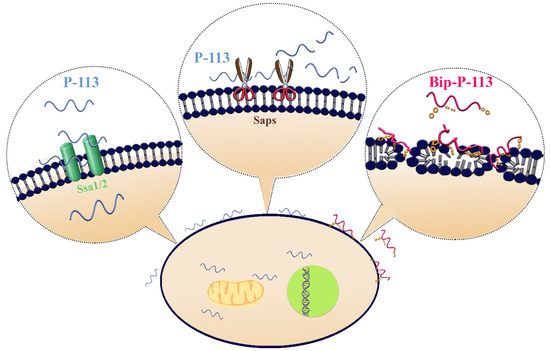
Figure 4. Model of the proposed interactions of P-113 and Bip-P-113 with Candida albicans. P-113 peptides recognize and bind to Ssa1/2 proteins and then translocate to the cytoplasm, leading to cell death. However, some of P-113 peptides are degraded by C. albicans secreted aspartic proteases (Saps). In contrast, Bip-P-113 accumulates in cell membrane and induces pore formation.
References
- Hui-Yuan Yu; Chih-Hsiung Tu; Bak-Sau Yip; Heng-Li Chen; Hsi-Tsung Cheng; Kuo-Chun Huang; Hsiu-Jung Lo; Jya-Wei Cheng; Easy Strategy To Increase Salt Resistance of Antimicrobial Peptides▿. Antimicrobial Agents and Chemotherapy 2011, 55, 4918-4921, 10.1128/AAC.00202-11.




