
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Khuram Razzaq | + 2110 word(s) | 2110 | 2021-02-10 04:56:04 | | | |
| 2 | Dean Liu | -1290 word(s) | 820 | 2021-02-18 08:37:44 | | |
Video Upload Options
The genome-editing tool, CRISPR-Cas9, reveals the functional features of several parts of the plant genome. Current developments in CRISPR, such as de novo meristem induction genome-engineering in dicots and temperature-tolerant LbCas12a/CRISPR, enable greater DNA insertion precision.
1. Introduction
In cells, CRISPR-Cas9 is a cheap, easy, fast, and effective system for gene knockout[1]. For effective genome engineering, CRISPR-Cas9 has been used in animals, plants, and bacteria[2][3][4][5]. Furthermore, CRISPR-Cas9 has been used for high-throughput screening of genes, gene knockout, chromosomal loci live-cell labeling, endogenous gene expression, and single-stranded RNA (ssRNA) edition. The application of CRISPR-Cas9 for studying the function of a gene has generated disease models. However, several queries and challenges need to be addressed. CRISPR-Cas9 will likely enhance our comprehension of disease activity and its management. For targeted genome engineering, detecting programmable nucleases that produce cuts in double-strands has radically changed molecular biology; ZFNs pioneered this success, with TALEN extending the genome modifying capacity[6]. Globally, CRISPR-Cas9 received recognition from researchers for its visible benefits over TALEN and ZFN[7], being its (1) ease of designing target, (2) ability to create mutations by inserting the guided RNA and Cas9 protein, and (3) multiplexing ability to target several genes at one time [8][9]. Figure 1 summarizes the principles of CRISPR-Cas9.
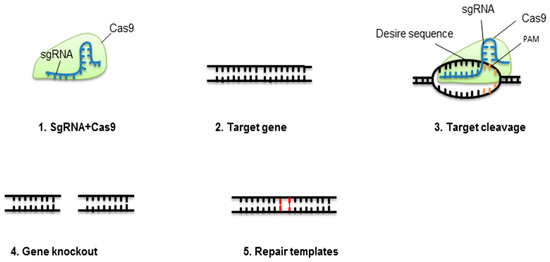
Figure 1. Concept of CRISPR-Cas9-mediated gene elimination. Single guide RNA (sgRNA) containing crRNA and tracrRNA fixes to Cas9 protein. This complex will break at a specific target of the double-stranded DNA molecule. The nonhomologous end-joining pathway (NHEJ) will repair the cleaved location.
2. CRISPR Technology
Due to its robust success, CRISPR-Cas9 is becoming a potential tool for genetically enhancing desirable crop traits, i.e., disease resistance, nutrient content, adaptation to multiple stresses, plant architecture, and yield. In some cases, a specific trait can be improved by negative regulatory gene knockout. Rice grain weight improved with gene modification of some QTL[10]. Maize grain yield under drought increased with genome engineering of the ARGOS8 locus[11]. In woody plants, CRISPR-Cas9 produced mutants in the first transgenic generation; this is significant as woody plant breeding is difficult due to their long lifespan[12][13]. Another study knocked out the OsGAN1 gene in rice and verified that it regulates root length and plant height[14]. Similarly, OsABCG26 gene knockout verified that this gene regulates pollen exine and anther cuticle, and OsTCD10 had a substantial role in chloroplasts of cold-stressed rice[15][16].
3. CRISPR-Cas9 Genome Editing to Biotic Stress Tolerance
Genome editing by CRISPR-Cas9 has been used effectively in several crops, including cotton, maize, rice, and wheat. However, most genome engineering studies have targeted biotic stresses, such as diseases. In wheat, the CRISPR-Cas9 method was used successfully to knock out all three EDR1 homologs to create plants (Taedr1) with increased tolerance to powdery mildew [17]. In Arabidopsis, the knockout of susceptible gene EDR1 increased resistance to powdery mildew[18]. Recessive resistance genes, eIF (eukaryotic translation initiation factor), have been detected in several dissimilar hosts, with eIF (iso) 4E and eIF4E genes used with CRISPR-Cas9 to form virus-resistant plants in Arabidopsis and cucumber, respectively[19][20]. CsLOB1 is a susceptible gene of the citrus canker (causative agent; Xanthomonascitri); CRISPR-Cas9 was used to edit this gene to develop resistant grapefruit plants [21][22]. Additionally, a negative resistance function MLO gene, responsible for powdery mildew susceptibility, was mutated successfully by Cas9 knockouts to enhance resistance against powdery mildew in tomato and wheat [23][24][25]. The application of CRISPR-Cas9 as an antivirus tool cleaved beet severe curly top virus, which decreased the viral infection[26][27]. The rice tungro spherical virus (RTSV), linked to the negatively controlled susceptible eIF4G gene, was eliminated using CRISPR-Cas9 to develop resistant rice varieties[28]. From CRISPR-Cas9, the loss of function VvWRKY52 gene produced resistance against Botrytis cinerea in grape (Vitis vinifera)[29]. Furthermore, CRISPR-Cas9 has been used to interrupt multiple virus genomes, including CLCuK0V, TYLCSV, and TYLCV[30]. For cucumber mosaic virus and tobacco mosaic virus, a technology to modify RNA virus genomes has been advanced from sgRNA and FnCas9. Hence, molecular immunity to RNA viruses was mediated by sgRNA/FnCas9 expression in Arabidopsis and tobacco[31]. CRISPR-Cas9 successfully targeted OsERF922 against blast fungus resistance in rice[32]. Plant ethylene-responsive factors (ERFs) can control tolerance against various stresses because they are involved in the ethylene (cytokinin) pathway[33]. When taken together, these reports deliver robust indications that CRISPR-Cas9 can enhance biotic stress resistance in plants (Table 2).
Table 2. CRISPR-Cas9 application for crop improvement.
| Species | Traits | Target Genes | Reference |
|---|---|---|---|
| Abiotic stresses | |||
| Rice | Improved resistance to arsenic stress | ARM1 | [34] |
| Depletion of Cd into grain | LCT1 | [35] | |
| Depletion of Cd into grain | Nramp5 | [36] | |
| Drought tolerance | SAPK2 | [37] | |
| Tomato | Drought tolerance | SIMAPK3 | [58] |
| Maize | Drought tolerance | ARGOS8 | [39] |
| Arabidopsis | Cold tolerance | CBF1 CBF2 | [40] |
| Biotic stresses | |||
| Arabidopsis | Resistance to turnip mosaic virus | eIF (iso)4E | [20] |
| Wheat | Improved resistance to powdery mildew | TaMLO | [25] |
| Improved resistance to powdery mildew | EDR1 | [17] | |
| Rice | Increased resistance to blast fungus | OsERF922 | [32] |
| Increased resistance to tungro spherical virus | eIF4G | [28] | |
| Barley | Improved resistance to fungal pathogens | MORC1 | [41] |
| Orange | Improved resistance to citrus canker | CsLOB1 | [42] |
| Tomato | Improved resistance to powdery mildew | Mlo1 | [24] |
| Anthocyanin biosynthesis | ANT1 | [43] | |
| Grape | Improved resistance to Botrytis cinerea | WRKY52 | [29] |
| Cucumber | Virus resistance | eIF4E | [19] |
References
- Khan, N.; Bano, A.; Rahman, M.A.; Rathinasabapathi, B.; Babar, M.A. UPLC-HRMS-based untargeted metabolic profiling reveals
- changes in chickpea (Cicer arietinum) metabolome following long-term drought stress. Plant Cell Environ. 2019, 42, 115.
- Feng, Z.; Zhang, B.; Ding,W.; Liu, X.; Yang, D.-L.;Wei, P.; Cao, F.; Zhu, S.; Zhang, F.; Mao, Y. Efficient genome editing in plants
- using a CRISPR/Cas system. Cell Res. 2013, 23, 1229.
- Jiang,W.; Zhou, H.; Bi, H.; Fromm, M.; Yang, B.;Weeks, D.P. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene
- modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 2013, 41, e188.
- Li, J.-F.; Norville, J.E.; Aach, J.; McCormack, M.; Zhang, D.; Bush, J.; Church, G.M.; Sheen, J. Multiplex and homologous
- recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol.
- 2013, 31, 688.
- Shan, Q.; Wang, Y.; Li, J.; Zhang, Y.; Chen, K.; Liang, Z.; Zhang, K.; Liu, J.; Xi, J.J.; Qiu, J.-L. Targeted genome modification of crop
- plants using a CRISPR-Cas system. Nat. Biotechnol. 2013, 31, 686.
- Chandrasegaran, S.; Carroll, D. Origins of programmable nucleases for genome engineering. J. Mol. Biol. 2016, 428, 963.
- Mao, Y.; Zhang, H.; Xu, N.; Zhang, B.; Gou, F.; Zhu, J.-K. Application of the CRISPR–Cas system for efficient genome engineering
- in plants. Mol. Plant 2013, 6, 2008.
- Ma, X.; Zhang, Q.; Zhu, Q.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.; Li, H.; Lin, Y. A robust CRISPR/Cas9 system for
- convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 2015, 8, 1274.
- Malzahn, A.; Lowder, L.; Qi, Y. Plant genome editing with TALEN and CRISPR. Cell Biosci. 2017, 7, 21.
- Xu, R.; Yang, Y.; Qin, R.; Li, H.; Qiu, C.; Li, L.; Wei, P.; Yang, J. Rapid improvement of grain weight via highly efficient
- CRISPR/Cas9-mediated multiplex genome editing in rice. J. Genet. Genom. Yi Chuanxuebao 2016, 43, 529.
- Shi, J.; Gao, H.;Wang, H.; Lafitte, H.R.; Archibald, R.L.; Yang, M.; Hakimi, S.M.; Mo, H.; Habben, J.E. ARGOS 8 variants generated
- by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol. J. 2017, 15, 207.
- Fan, D.; Liu, T.; Li, C.; Jiao, B.; Li, S.; Hou, Y.; Luo, K. Efficient CRISPR/Cas9-mediated targeted mutagenesis in Populus in the
- first generation. Sci. Rep. 2015, 5, 12217.
- Tsai, C.-J.; Xue, L.-J. CRISPRing into the woods. GM Crop. Food 2015, 6, 206.
- Ma, X.; Feng, F.; Wei, H.; Mei, H.; Xu, K.; Chen, S.; Li, T.; Liang, X.; Liu, H.; Luo, L. Genome-wide association study for plant
- height and grain yield in rice under contrasting moisture regimes. Front. Plant Sci. 2016, 7, 1801.
- Chang, Z.; Chen, Z.; Yan,W.; Xie, G.; Lu, J.;Wang, N.; Lu, Q.; Yao, N.; Yang, G.; Xia, J. An ABC transporter, OsABCG26, is required
- for anther cuticle and pollen exine formation and pollen-pistil interactions in rice. Plant Sci. 2016, 253, 21.
- Wu, L.; Wu, J.; Liu, Y.; Gong, X.; Xu, J.; Lin, D.; Dong, Y. The rice pentatricopeptide repeat gene TCD10 is needed for chloroplast
- development under cold stress. Rice 2016, 9.
- Zhang, Y.; Bai, Y.;Wu, G.; Zou, S.; Chen, Y.; Gao, C.; Tang, D. Simultaneous modification of three homoeologs of Ta EDR 1 by
- genome editing enhances powdery mildew resistance in wheat. Plant J. 2017, 91, 714.
- Frye, C.A.; Tang, D.; Innes, R.W. Negative regulation of defense responses in plants by a conserved MAPKK kinase. Proc. Natl.
- Acad. Sci. USA 2001, 98, 373.
- Chandrasekaran, J.; Brumin, M.; Wolf, D.; Leibman, D.; Klap, C.; Pearlsman, M.; Sherman, A.; Arazi, T.; Gal-On, A. Development
- of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol. Plant Pathol. 2016, 17, 1140.
- Pyott, D.E.; Sheehan, E.; Molnar, A. Engineering of CRISPR/Cas9-mediated potyvirus resistance in transgene-free Arabidopsis
- plants. Mol. Plant Pathol. 2016, 17, 1276.
- Hu, Y.; Zhang, J.; Jia, H.; Sosso, D.; Li, T.; Frommer,W.B.; Yang, B.; White, F.F.;Wang, N.; Jones, J.B. Lateral organ boundaries 1 is a
- disease susceptibility gene for citrus bacterial canker disease. Proc. Natl. Acad. Sci. USA 2014, 111, E521.
- Jia, H.; Zhang, Y.; Orbovi´c, V.; Xu, J.; White, F.F.; Jones, J.B.; Wang, N. Genome editing of the disease susceptibility gene Cs LOB 1
- in citrus confers resistance to citrus canker. Plant Biotechnol. J. 2017, 15, 817.
- Humphry, M.; Consonni, C.; Panstruga, R. mlo-based powdery mildew immunity: Silver bullet or simply non-host resistance?
- Mol. Plant Pathol. 2006, 7, 605.
- Nekrasov, V.; Wang, C.; Win, J.; Lanz, C.; Weigel, D.; Kamoun, S. Rapid generation of a transgene-free powdery mildew resistant
- tomato by genome deletion. Sci. Rep. 2017, 7.
- Wang, Y.; Cheng, X.; Shan, Q.; Zhang, Y.; Liu, J.; Gao, C.; Qiu, J.-L. Simultaneous editing of three homoeoalleles in hexaploid
- bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 2014, 32, 947.
- Chaparro-Garcia, A.; Kamoun, S.; Nekrasov, V. Boosting plant immunity with CRISPR/Cas. Genome Biol. 2015, 16, 254.
- Ji, X.; Zhang, H.; Zhang, Y.; Wang, Y.; Gao, C. Establishing a CRISPR-Cas-like immune system conferring DNA virus resistance in
- plants. Nat. Plants 2015, 1, 15144.
- Macovei, A.; Sevilla, N.R.; Cantos, C.; Jonson, G.B.; Slamet-Loedin, I.; Cˇ ermák, T.; Voytas, D.F.; Choi, I.R.; Chadha-Mohanty, P.
- Novel alleles of rice eIF4G generated by CRISPR/Cas9-targeted mutagenesis confer resistance to Rice tungro spherical virus.
- Plant Biotechnol. J. 2018, 16, 1918.
- Wang, X.; Tu, M.;Wang, D.; Liu, J.; Li, Y.; Li, Z.;Wang, Y.;Wang, X. CRISPR/Cas9-mediated efficient targeted mutagenesis in
- grape in the first generation. Plant Biotechnol. J. 2018, 16, 844.
- Zaidi, S.S.-E.-A.; Tashkandi, M.; Mansoor, S.; Mahfouz, M.M. Engineering plant immunity: Using CRISPR/Cas9 to generate virus
- resistance. Front. Plant Sci. 2016, 7, 1673.
- Zhang, T.; Zheng, Q.; Yi, X.; An, H.; Zhao, Y.; Ma, S.; Zhou, G. Establishing RNA virus resistance in plants by harnessing CRISPR
- immune system. Plant Biotechnol. J. 2018, 16, 1415.
- Wang, F.; Wang, C.; Liu, P.; Lei, C.; Hao, W.; Gao, Y.; Liu, Y.-G.; Zhao, K. Enhanced rice blast resistance by CRISPR/Cas9-targeted
- mutagenesis of the ERF transcription factor gene OsERF922. PLoS ONE 2016, 11, e0154027.
- Jung, J.;Won, S.Y.; Suh, S.C.; Kim, H.;Wing, R.; Jeong, Y.; Hwang, I.; Kim, M. The barley ERF-type transcription factor HvRAF
- confers enhanced pathogen resistance and salt tolerance in Arabidopsis. Planta 2007, 225, 575.
- Wang, F.-Z.; Chen, M.-X.; Yu, L.-J.; Xie, L.-J.; Yuan, L.-B.; Qi, H.; Xiao, M.; Guo, W.; Chen, Z.; Yi, K. OsARM1, an R2R3 MYB
- transcription factor, is involved in regulation of the response to arsenic stress in rice. Front. Plant Sci. 2017, 8, 1868.
- Lu, H.P.; Liu, S.M.; Xu, S.L.; Chen, W.Y.; Zhou, X.; Tan, Y.Y.; Huang, J.Z.; Shu, Q.Y. CRISPR-S: An active interference element for a
- rapid and inexpensive selection of genome-edited, transgene-free rice plants. Plant Biotechnol. J. 2017, 15, 1371.
- Tang, L.; Mao, B.; Li, Y.; Lv, Q.; Zhang, L.; Chen, C.; He, H.; Wang, W.; Zeng, X.; Shao, Y. Knockout of OsNramp5 using the
- CRISPR/Cas9 system produces low Cd-accumulating indica rice without compromising yield. Sci. Rep. 2017, 7.
- Lou, D.; Wang, H.; Liang, G.; Yu, D. OsSAPK2 confers abscisic acid sensitivity and tolerance to drought stress in rice. Front. Plant
- Sci. 2017, 8, 993.
- Wang, L.; Chen, L.; Li, R.; Zhao, R.; Yang, M.; Sheng, J.; Shen, L. Reduced drought tolerance by CRISPR/Cas9-mediated SlMAPK3
- mutagenesis in tomato plants. J. Agric. Food Chem. 2017, 65, 8674.
- Hirai, M.Y.; Sugiyama, K.; Sawada, Y.; Tohge, T.; Obayashi, T.; Suzuki, A.; Goda, H. Omics based identification of Arabidopsis Myb
- transcription factors regulating aliphatic glucosinolate biosynthesis. Proc. Natl. Acad. Sci. USA 2007, 104, 6478.
- Chen, H.-J.; Su, C.-T.; Lin, C.-H.; Huang, G.-J.; Lin, Y.-H. Expression of sweet potato cysteine protease SPCP2 altered developmental
- characteristics and stress responses in transgenic Arabidopsis plants. J. Plant Physiol. 2010, 167, 838.
- Kumar, N.; Galli, M.; Ordon, J.; Stuttmann, J.; Kogel, K.H.; Imani, J. Further analysis of barley MORC 1 using a highly efficient
- RNA-guided Cas9 gene-editing system. Plant Biotechnol. J. 2018, 16, 1892.
- Peng, A.; Chen, S.; Lei, T.; Xu, L.; He, Y.; Wu, L.; Yao, L.; Zou, X. Engineering canker-resistant plants through CRISPR/Cas9-
- targeted editing of the susceptibility gene Cs LOB 1 promoter in citrus. Plant Biotechnol. J. 2017, 15, 1509.
- Cˇ ermák, T.; Baltes, N.J.; Cˇ egan, R.; Zhang, Y.; Voytas, D.F. High-frequency, precise modification of the tomato genome. Genome
- Biol. 2015, 16, 232.




