| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Katarzyna Retzer | + 1833 word(s) | 1833 | 2021-01-21 03:46:26 | | | |
| 2 | Katarzyna Retzer | -21 word(s) | 1812 | 2021-02-01 23:04:10 | | | | |
| 3 | Bruce Ren | Meta information modification | 1812 | 2021-02-02 04:09:46 | | |
Video Upload Options
Plant growth and productivity are orchestrated by a network of signaling cascades involved in balancing responses to perceived environmental changes with resource availability. Vascular plants are divided into the shoot, an aboveground organ where sugar is synthesized, and the underground located root. Continuous growth requires the generation of energy in the form of carbohydrates in the leaves upon photosynthesis and uptake of nutrients and water through root hairs. Root hair outgrowth depends on the overall condition of the plant and its energy level must be high enough to maintain root growth. TARGET OF RAPAMYCIN (TOR)-mediated signaling cascades serve as a hub to evaluate which resources are needed to respond to external stimuli and which are available to maintain proper plant growth. Root hair formation further requires appropriate distribution of the phytohormone auxin, which primes root hair cell fate and triggers root hair elongation. While TOR activity partially depends on auxin signaling, auxin distribution to the root tip and the redistribution towards the root hair zone are, on the other hand, dependent on TOR activity. Together, both signaling cascades are interwoven to orchestrate proper root hair outgrowth to ensure efficient plant nutrition.
1. Introduction
Plant growth, and, more critically, its adaptation to ever-changing environmental conditions, is maintained by the ability of plants to perceive exogenous signals, to evaluate the availability of resources, and to initiate signaling cascades orchestrating changes in growth rate and direction [1][2][3]. The most crucial aspect of plant fitness is to keep the potential of balancing the amount of cell division in meristematic regions and elongation processes and differentiation to mature cells, which depends on environmental conditions and available resources [4]. Efficient growth requires the generation of energy in form of sugars in the leaves of the plant upon photosynthesis, which is followed by the biosynthesis of the phytohormone and growth regulator auxin [5][6]. Furthermore, the plant needs to regulate nutrient and water uptake through the root from the soil [7][8]. To enhance uptake efficiency, the root has the ability to grow tubular structures from specialized epidermis cells, the root hairs [7][8]. Root hair cell priming and outgrowth are highly dependent on proper auxin distribution and abundance, whereby both are dependent on environmental conditions and the overall condition of the plant [8][9][10][11]. Distribution of the phytohormone auxin through the plant creates auxin gradients, which orchestrate the activity of meristems, adaptational growth via cell expansion, and differentiation of specialized cells [12][13][14][15]. PIN-FORMED (PIN) proteins, auxin efflux carriers, ensure on-point and polar distribution of the phytohormone, whereby auxin signaling underpins the fine-tuning of various cellular processes involved in plant growth adaptation under changing environmental conditions [16][17][18]. The role of the root-specific PIN family member PIN2 during root hair growth adaptation in response to environmental conditions will be discussed in particular in the scope of this review. The establishment of auxin gradients is crucial for plant growth from the first asymmetric cell division of the zygote [19][20] and tissue organization [21] through fine-tuning cell proliferation and cell elongation [22][23][24][25], which in the end ensures that the shoot will grow above the soil, whereas the root anchors the plant in the soil. Shoot growth is primarily oriented towards the light (positive phototropic, negative gravitropic) to ensure the most efficient exposure to the sun to harvest energy, which is converted to sugar and powers plant growth [26]. In contrast, the root grows along the gravity vector (positive gravitropic, negative phototropic) to anchor it in the soil, where it absorbs nutrients and water. Auxin biosynthesis correlates with the sugar levels produced depending on light quality and quantity perceived by the leaves and transported to the roots [27][28]. The available auxin orchestrates the establishment of root system architecture including, among others, the length of the primary root, the number of lateral roots, and the outgrowth of root hairs [28][29][30]. Auxin is actively transported towards the root meristem, where it modulates root growth depending on the requirements in a balance between environmental stimuli and available resources, such as carbohydrates, minerals, and water [31]. The information about the energy status and available resources of the plant are sensed by the TARGET OF RAPAMYCIN (TOR) complex, which then activates cellular responses upstream of root hair outgrowth (Figure 1).
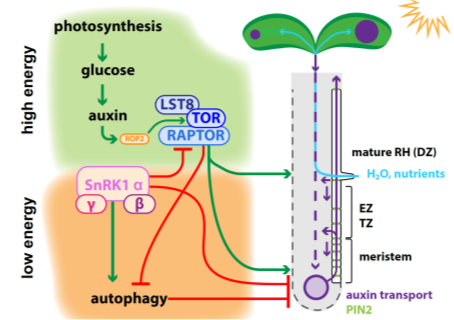
Figure 1. Energy status and resource uptake are orchestrated by an interplay of TARGET OF RAPAMYCIN (TOR) and SNF1-RELATED KINASE 1 (SnRK1) complexes, which antagonistically modulate root and root hair growth. High energy status results from sugar synthesis derived from photosynthesis in the leaves, which is followed by elevated auxin biosynthesis. Auxin signaling enhances RHO-RELATED PROTEIN FROM PLANTS 2 (ROP2) , and ROP2 binds TOR and promotes its phosphorylation. This results in the activation of the TOR complex, which further consists of the substrate-presenting subunit REGULATORY-ASSOCIATED PROTEIN OF TOR (RAPTOR) and the complex stabilizer LETHAL WITH SEC THIRTEEN 8 (LST8). TOR signaling positively correlates with root meristem activity and therefore with root length. TOR stabilizes the auxin efflux carrier PIN2 by phosphorylation at the plasma membrane, and thereby modulates shootward auxin transport. Auxin levels in the meristem enhance the cell proliferation rate and are involved in root hair cell priming . Furthermore, efficient auxin delivery and signaling trigger root hair elongation, which is responsible for root surface enlargement, allowing enhanced uptake of water and nutrients to ensure plant growth and productivity. Under high energy levels, TOR signaling inhibits autophagy, which would trigger the engulfment and recycling of existing plant material to gain resources, and this is accompanied by the end of root growth. When the energy levels in the root drop, reduced sugar availability is sensed over the SNF1-RELATED KINASE 1 (SnRK1) complex, which balances between TOR complex deactivation and autophagy activation. SnRK1 complex activity results in RAPTOR deactivation by phosphorylation, and the downregulation of auxin efflux transporters involved in auxin transport from the shoot to the root, followed by reduced root meristem activity. EZ = elongation zone; TZ = transition zone; DZ = differentiation zone.
2. Plant Growth is Dynamic and Orchestrated by Interwoven Signaling Cascades Orchestrated by TOR
Successful plant growth and productivity are highly dependent on the establishment and maintenance of polarity on the tissue, cell, and subcellular levels[32][33]. Plants evolved a fine-tuned network of intracellular processes to adapt to environmental changes, leading to fast adaptation responses and manifesting in the structural reorganization of plant architecture to ensure proper nutrient distribution and growth [34]. Manifold signaling cascades are simultaneously active along the plant body, which implement exogenous signals and information of available resources and the energy status of the shoot and the root. TARGET OF RAPAMYCIN (TOR) is a core hub among signaling cascades to balance cellular responses upon hormonal and nutrient signaling, including root meristem activity and root hair outgrowth [35][36][37][38].
TOR is a Ser/Thr protein kinase, structurally and functionally conserved among all eukaryotes, and acts as a master regulator of transition between cell proliferation and elongation. Furthermore, it inhibits autophagy in nutrient-rich conditions [38] (Figure 1). TOR associates with two other proteins in plants, REGULATORY-ASSOCIATED PROTEIN OF TOR (RAPTOR), which presents substrate proteins to TOR, and LETHAL WITH SEC THIRTEEN 8 (LST8), responsible for stabilizing the complex [39] (Figure 1). TOR signaling networks are crucial to adjusting metabolism rates during plant growth [40][41][42][43]. As a crucial part of the glucose signaling network, the TOR complex modulates the activity of transcription factors, such as E2Fα, to enhance root meristem activity [44]. TOR signaling positively regulates the energy-demanding protein translation machinery, but is inhibited when the energy status of the plant drops [45]. In elongated starvation, the plant needs to shut down de novo protein synthesis, and furthermore, the TOR inhibitory effect on autophagy is reversed in Arabidopsis thaliana [46], as well in the green alga Chlamydomonas reinhardtii [47][48][49]. When the available resources do not cover the plant’s energy demand anymore, cell material undergoes recycling [50][51]. The drop in energy levels of the plant is sensed by the SNF1-RELATED KINASE 1 (SnRK1) complex, which serves as a plant’s metabolic sensor to keep track of carbohydrate level changes [52]. The heterotrimeric SnRK1 complex is composed of one catalytic and two regulatory subunits (Figure 1). Three isoforms of the catalytic subunit are known to be encoded in the Arabidopsis thaliana genome, but only SnRK1α1/AKIN10/KIN10 is responsible for most of the SnRK1 activity[53]. Under nutrient-rich conditions, SnRK1 activity is kept low, but the reduction of available resources leads to the transcriptional upregulation of SnRK1 levels [53]. SnRK1 complex activity further results in the activation of autophagy to regain resources [52]. Recently, it has been shown that the overexpression of SnRK1α1 leads to the downregulation of the TOR/S6K/RPS6 phosphorylation cascade, while SnRK1α1 suppression causes the opposite effect The TOR/S6K/RPS6 phosphorylation cascade is supposed to be involved in high energy-demanding polysome abundance and protein translation activity as well as stress responses [54][55]. It has been shown that SnRK1α1 physically interacts with RAPTOR and is able to phosphorylate the TOR complex, thereby potentially downregulating its activity (Figure 1). When energy levels rise again, as is the case of elevated photosynthetic activity, rising sugar levels are followed by an enhanced auxin biosynthesis, whereas elevated auxin levels are connected with the activation of TOR by phosphorylation (Figure 1). Besides sugar levels, the availability of amino acids underpins TOR activity, like tryptophan, which is an important precursor for the biosynthesis of auxins [54][55][56][57][58][59]. TOR signaling and auxin abundance and distribution are tightly interconnected, as auxin levels decrease upon treatment with TOR inhibitors in the root [60]. Among other amino acids, the abundance of cysteine is critical for plant productivity and is also highly dependent on sufficient sulfur uptake [61]. Sulfur depletion is transduced over TOR signaling, resulting in the downregulation of glucose metabolism and the upregulation of autophagy, followed by reduced meristem activity [61]. Sulfate uptake is tightly coupled with carbohydrate and nitrogen metabolism [61], and TOR signaling is thereby a centerpiece of the integration and coordination of light, hormone, and metabolism sensing and signaling upstream of root and shoot architecture adaptation. TOR activity is also regulated by phosphorylation, which, as described above, correlates with auxin abundance in the presence of glucose [62]. Enhanced TOR activity is coupled to increased plant growth rate, fitness, and size [63]. In the root, shoot-derived glucose and auxin signaling together activate TOR signaling pathways, leading to the upregulation of cell proliferation in the meristem, which further positively correlates with root hair growth [64]. Estradiol-inducible tor-es mutant and raptor1b roots are impaired in glucose-activated root hair elongation [65]. Furthermore, TOR inhibitor treatments also suggest a link to auxin biosynthesis and signaling in root hair development . Auxin activates the TOR complex over RHO-RELATED PROTEIN FROM PLANTS 2 (ROP2) action [66] (Figure 1). To date, the interaction between auxin, ROP2, and TOR upstream of TOR signaling on the whole plant level has been well studied , but how ROP2 activity and polar auxin transport are precisely regulated during root hair growth initiation depending on TOR signaling must be still determined [67]. Auxin distribution to the root tip and the redistribution towards the root hair zone are, on the other hand, dependent on TOR activity, showing how exogenous and hormonal signals influence each other .
References
- Van Gelderen, K.; Kang, C.; Pierik, R. Light Signaling, Root Development, and Plasticity. Plant Physiol. 2018, doi:10.1104/pp.17.01079.
- Barrada, A.; Montané, M.H.; Robaglia, C.; Menand, B. Spatial Regulation of Root Growth: Placing the Plant TOR Pathway in a Developmental Perspective. Int. J. Mol. Sci. 2015, doi:10.3390/ijms160819671.
- Retzer, K.; Korbei, B.; Luschnig, C. Auxin and Tropisms. In Auxin and Its Role in Plant Development; Springer: Berlin, Germa-ny, 2014.
- Calleja-Cabrera, J.; Boter, M.; Oñate-Sánchez, L.; Pernas, M. Root Growth Adaptation to Climate Change in Crops. Front. Plant Sci. 2020, doi:10.3389/fpls.2020.00544.
- Kircher, S.; Schopfer, P. Photosynthetic Sucrose Acts as Cotyledon-Derived Long-Distance Signal to Control Root Growth during Early Seedling Development in Arabidopsis. Proc. Natl. Acad. Sci. USA 2012, doi:10.1073/pnas.1203746109.
- Bennett, T.; Leyser, O. The Auxin Question: A Philosophical Overview. In Auxin and Its Role in Plant Development; Springer: Berlin, Germany, 2014.
- Dolan, L.; Duckett, C.M.; Grierson, C.; Linstead, P.; Schneider, K.; Lawson, E.; Dean, C.; Poethig, S.; Roberts, K. Clonal Rela-tionships and Cell Patterning in the Root Epidermis of Arabidopsis. Development 1994, 120, 2465–2474.
- Grierson, C.; Nielsen, E.; Ketelaarc, T.; Schiefelbein, J. Root Hairs. Arab. Book 2014, 12, e0172–e0172, doi:10.1199/tab.0172.
- Jones, A.R.; Kramer, E.M.; Knox, K.; Swarup, R.; Bennett, M.J.; Lazarus, C.M.; Leyser, H.M.O.; Grierson, C.S. Auxin Transport through Non-Hair Cells Sustains Root-Hair Development. Nat. Cell Biol. 2009, doi:10.1038/ncb1815.
- Leyser, O. Auxin Signaling. Plant Physiol. 2018, doi:10.1104/pp.17.00765.
- Wu, Y.; Shi, L.; Li, L.; Fu, L.; Liu, Y.; Xiong, Y.; Sheen, J. Integration of Nutrient, Energy, Light, and Hormone Signalling via TOR in Plants. J. Exp. Bot. 2019, doi:10.1093/jxb/erz028.
- Sauer, M.; Kleine-Vehn, J. PIN-FORMED and PIN-LIKES Auxin Transport Facilitators. Development 2019, doi:10.1242/dev.168088.
- Muroyama, A.; Bergmann, D. Plant Cell Polarity: Creating Diversity from inside the Box. Annu. Rev. Cell Dev. Biol. 2019, doi:10.1146/annurev-cellbio-100818-125211.
- Adamowski, M.; Friml, J. PIN-Dependent Auxin Transport: Action, Regulation, and Evolution. Plant Cell 2015, doi:10.1105/tpc.114.134874.
- Qi, J.; Greb, T. Cell Polarity in Plants: The Yin and Yang of Cellular Functions. Curr. Opin. Plant Biol. 2017, doi:10.1016/j.pbi.2016.11.015.
- Ötvös, K.; Marconi, M.; Vega, A.; O’Brien, J.; Johnson, A.; Abualia, R.; Antonielli, L.; Montesinos, J.C.; Zhang, Y.; Tan, S.; et al. Modulation of Root Growth by Nutrient-Defined Fine-Tuning of Polar Auxin Transport. bioRxiv 2020, doi:10.1101/2020.06.19.160994.
- Band, L.R.; Wells, D.M.; Larrieu, A.; Sun, J.; Middleton, A.M.; French, A.P.; Brunoud, G.; Sato, E.M.; Wilson, M.H.; Peŕet, B.; et al. Root Gravitropism Is Regulated by a Transient Lateral Auxin Gradient Controlled by a Tipping-Point Mechanism. Proc. Natl. Acad. Sci. USA 2012, doi:10.1073/pnas.1201498109.
- Park, E.; Nebenführ, A. Cytoskeleton and Root Hair Growth. Plant Cytoskelet. 2011, doi:10.1007/978-1-4419-0987-9_12.
- Friml, J.; Vieten, A.; Sauer, M.; Weijers, D.; Schwarz, H.; Hamann, T.; Offringa, R.; Jürgens, G. Efflux-Dependent Auxin Gra-dients Establish the Apical-Basal Axis of Arabidopsis. Nature 2003, doi:10.1038/nature02085.
- Smit, M.E.; Weijers, D. The Role of Auxin Signaling in Early Embryo Pattern Formation. Curr. Opin. Plant Biol. 2015, doi:10.1016/j.pbi.2015.10.001.
- Benková, E.; Michniewicz, M.; Sauer, M.; Teichmann, T.; Seifertová, D.; Jürgens, G.; Friml, J. Local, Efflux-Dependent Auxin Gradients as a Common Module for Plant Organ Formation. Cell 2003, doi:10.1016/S0092-8674(03)00924-3.
- Armengot, L.; Marquès-Bueno, M.M.; Jaillais, Y. Regulation of Polar Auxin Transport by Protein and Lipid Kinases. J. Exp. Bot. 2016, doi:10.1093/jxb/erw216.
- Fendrych, M.; Akhmanova, M.; Merrin, J.; Glanc, M.; Hagihara, S.; Takahashi, K.; Uchida, N.; Torii, K.U.; Friml, J. Rapid and Reversible Root Growth Inhibition by TIR1 Auxin Signalling. Nat. Plants 2018, doi:10.1038/s41477-018-0190-1.
- Gallei, M.; Luschnig, C.; Friml, J. Auxin Signalling in Growth: Schrödinger’s Cat out of the Bag. Curr. Opin. Plant Biol. 2020, doi:10.1016/j.pbi.2019.10.003.
- Semeradova, H.; Montesinos, J.C.; Benkova, E. All Roads Lead to Auxin: Post-Translational Regulation of Auxin Transport by Multiple Hormonal Pathways. Plant Commun. 2020, doi:10.1016/j.xplc.2020.100048.
- van Gelderen, K.; Kang, C.; Paalman, R.; Keuskamp, D.; Hayes, S.; Pierik, R. Far-Red Light Detection in the Shoot Regulates Lateral Root Development through the HY5 Transcription Factor. Plant Cell 2018, doi:10.1105/tpc.17.00771.
- Sairanen, I.; Novák, O.; Pěnčík, A.; Ikeda, Y.; Jones, B.; Sandberg, G.; Ljung, K. Soluble Carbohydrates Regulate Auxin Bio-synthesis via PIF Proteins in Arabidopsis. Plant Cell 2013, doi:10.1105/tpc.112.104794.
- Pierik, R.; Testerink, C. The Art of Being Flexible: How to Escape from Shade, Salt, And Drought1. Plant Physiol. 2014, doi:10.1104/pp.114.239160.
- de Smet, I. Lateral Root Initiation: One Step at a Time. New Phytol. 2012, doi:10.1111/j.1469-8137.2011.03996.x.
- Orman-Ligeza, B.; Parizot, B.; Gantet, P.P.; Beeckman, T.; Bennett, M.J.; Draye, X. Post-Embryonic Root Organogenesis in Cereals: Branching out from Model Plants. Trends Plant Sci. 2013, doi:10.1016/j.tplants.2013.04.010.
- Habets, M.E.J.; Offringa, R. PIN-Driven Polar Auxin Transport in Plant Developmental Plasticity: A Key Target for Envi-ronmental and Endogenous Signals. New Phytol. 2014, 362–377, doi:10.1111/nph.12831.
- Rakusová, H.; Fendrych, M.; Friml, J. Intracellular Trafficking and PIN-Mediated Cell Polarity during Tropic Responses in Plants. Curr. Opin. Plant Biol. 2015, 116–123, doi:10.1016/j.pbi.2014.12.002.
- Wallner, E.-S. The Value of Asymmetry: How Polarity Proteins Determine Plant Growth and Morphology. J. Exp. Bot. 2020, 71, 5733–5739, doi:10.1093/jxb/eraa329.
- Gupta, A.; Rico-Medina, A.; Caño-Delgado, A.I. The Physiology of Plant Responses to Drought. Science 2020, doi:10.1126/science.aaz7614.
- Bögre L., Henriques R., Magyar Z., TOR tour to auxin. EMBO J, 2013, https://doi.org/10.1038/emboj.2013.69..
- Yokawa, K.; Baluška, F. The TOR Complex: An Emergency Switch for Root Behavior. Plant Cell Physiol. 2016, doi:10.1093/pcp/pcv191.
- Ryabova, L.A.; Robaglia, C.; Meyer, C. Target of Rapamycin Kinase: Central Regulatory Hub for Plant Growth and Metabo-lism. J. Exp. Bot. 2019, doi:10.1093/jxb/erz108.
- Zhao, Y.; Wang, X.Q. The Hot Issue: TOR Signalling Network in Plants. Funct. Plant Biol. 2020, doi:10.1071/FP20071.
- Soto-Burgos, J.; Bassham, D.C. SnRK1 Activates Autophagy via the TOR Signaling Pathway in Arabidopsis Thaliana. PLoS ONE 2017, doi:10.1371/journal.pone.0182591.
- Roustan, V.; Jain, A.; Teige, M.; Ebersberger, I.; Weckwerth, W. An Evolutionary Perspective of AMPK-TOR Signaling in the Three Domains of Life. J. Exp. Bot. 2016, doi:10.1093/jxb/erw211.
- Mair, A.; Pedrotti, L.; Wurzinger, B.; Anrather, D.; Simeunovic, A.; Weiste, C.; Valerio, C.; Dietrich, K.; Kirchler, T.; Nägele, T.; et al. SnRK1-Triggered Switch of BZIP63 Dimerization Mediates the Low-Energy Response in Plants. eLife 2015, doi:10.7554/eLife.05828.
- Xiong, Y.; Sheen, J. Novel Links in the Plant TOR Kinase Signaling Network. Curr. Opin. Plant Biol. 2015, doi:10.1016/j.pbi.2015.09.006.
- Shi, L.; Wu, Y.; Sheen, J. TOR Signaling in Plants: Conservation and Innovation. Development 2018, doi:10.1242/dev.160887.
- Xiong, Y.; McCormack, M.; Li, L.; Hall, Q.; Xiang, C.; Sheen, J. Glucose-TOR Signalling Reprograms the Transcriptome and Activates Meristems. Nature 2013, doi:10.1038/nature12030.
- Nukarinen, E.; Ngele, T.; Pedrotti, L.; Wurzinger, B.; Mair, A.; Landgraf, R.; Börnke, F.; Hanson, J.; Teige, M.; Baena-Gonzalez, E.; et al. Quantitative Phosphoproteomics Reveals the Role of the AMPK Plant Ortholog SnRK1 as a Meta-bolic Master Regulator under Energy Deprivation. Sci. Rep. 2016, doi:10.1038/srep31697.
- Liu, Y.; Bassham, D.C. TOR Is a Negative Regulator of Autophagy in Arabidopsis Thaliana. PLoS ONE 2010, doi:10.1371/journal.pone.0011883.
- Pérez-Pérez, M.E.; Florencio, F.J.; Crespo, J.L. Inhibition of Target of Rapamycin Signaling and Stress Activate Autophagy in Chlamydomonas Reinhardtii. Plant Physiol. 2010, doi:10.1104/pp.109.152520.
- Roustan, V.; Bakhtiari, S.; Roustan, P.J.; Weckwerth, W. Quantitative in Vivo Phosphoproteomics Reveals Reversible Signal-ing Processes during Nitrogen Starvation and Recovery in the Biofuel Model Organism Chlamydomonas Reinhardtii. Bio-technol. Biofuels 2017, doi:10.1186/s13068-017-0949-z.
- Roustan, V.; Weckwerth, W. Quantitative Phosphoproteomic and System-Level Analysis of TOR Inhibition Unravel Distinct Organellar Acclimation in Chlamydomonas Reinhardtii. Front. Plant Sci. 2018, doi:10.3389/fpls.2018.01590.
- Batoko, H.; Dagdas, Y.; Baluska, F.; Sirko, A. Understanding and Exploiting Autophagy Signaling in Plants. Essays Biochem. 2017, doi:10.1042/EBC20170034.
- Masclaux-Daubresse, C. Autophagy Controls Carbon, Nitrogen, and Redox Homeostasis in Plants. Autophagy 2016, doi:10.4161/auto.36261.
- Margalha, L.; Confraria, A.; Baena-González, E. SnRK1 and TOR: Modulating Growth–Defense Trade-Offs in Plant Stress Responses. J. Exp. Bot. 2019, doi:10.1093/jxb/erz066.
- Margalha, L.; Valerio, C.; Baena-González, E. Plant SnRK1 Kinases: Structure, Regulation, and Function. Act. Protein Kinase 2016, doi:10.1007/978-3-319-43589-3_17.
- Dobrenel, T.; Mancera-Martínez, E.; Forzani, C.; Azzopardi, M.; Davanture, M.; Moreau, M.; Schepetilnikov, M.; Chicher, J.; Langella, O.; Zivy, M.; et al. The Arabidopsis TOR Kinase Specifically Regulates the Expression of Nuclear Genes Coding for Plastidic Ribosomal Proteins and the Phosphorylation of the Cytosolic Ribosomal Protein S6. Front. Plant Sci. 2016, doi:10.3389/fpls.2016.01611.
- Mahfouz, M.M.; Kim, S.; Delauney, A.J.; Verma, D.P.S. Arabidopsis TARGET of RAPAMYCIN Interacts with RAPTOR, Which Regulates the Activity of S6 Kinase in Response to Osmotic Stress Signals. Plant Cell 2006, doi:10.1105/tpc.105.035931.
- Schepetilnikov, M.; Dimitrova, M.; Mancera-Martínez, E.; Geldreich, A.; Keller, M.; Ryabova, L.A. TOR and S6K1 Promote Translation Reinitiation of UORF-Containing MRNAs via Phosphorylation of EIF3h. EMBO J. 2013, doi:10.1038/emboj.2013.61.
- Ljung, K. Auxin Metabolism and Homeostasis during Plant Development. Development 2013, doi:10.1242/dev.086363.
- Simon, S.; Petrášek, J. Why Plants Need More than One Type of Auxin. Plant Sci. 2011, doi:10.1016/j.plantsci.2010.12.007.
- Morffy, N.; Strader, L.C. Old Town Roads: Routes of Auxin Biosynthesis across Kingdoms. Curr. Opin. Plant Biol. 2020, doi:10.1016/j.pbi.2020.02.002.
- Deng, K.; Yu, L.; Zheng, X.; Zhang, K.; Wang, W.; Dong, P.; Zhang, J.; Ren, M. Target of Rapamycin Is a Key Player for Auxin Signaling Transduction in Arabidopsis. Front. Plant Sci. 2016, doi:10.3389/fpls.2016.00291.
- Dong, Y.; Silbermann, M.; Speiser, A.; Forieri, I.; Linster, E.; Poschet, G.; Allboje Samami, A.; Wanatabe, M.; Sticht, C.; Teleman, A.A.; et al. Sulfur Availability Regulates Plant Growth via Glucose-TOR Signaling. Nat. Commun. 2017, doi:10.1038/s41467-017-01224-w.
- Schepetilnikov, M.; Ryabova, L.A. Auxin Signaling in Regulation of Plant Translation Reinitiation. Front. Plant Sci. 2017, doi:10.3389/fpls.2017.01014.
- Deprost, D.; Yao, L.; Sormani, R.; Moreau, M.; Leterreux, G.; Bedu, M.; Robaglia, C.; Meyer, C. The Arabidopsis TOR Kinase Links Plant Growth, Yield, Stress Resistance and MRNA Translation. EMBO Rep. 2007, doi:10.1038/sj.embor.7401043.
- Mishra, B.S.; Singh, M.; Aggrawal, P.; Laxmi, A. Glucose and Auxin Signaling Interaction in Controlling Arabidopsis Thali-ana Seedlings Root Growth and Development. PLoS ONE 2009, doi:10.1371/journal.pone.0004502.
- Salem, M.A.; Li, Y.; Wiszniewski, A.; Giavalisco, P. Regulatory-Associated Protein of TOR (RAPTOR) Alters the Hormonal and Metabolic Composition of Arabidopsis Seeds, Controlling Seed Morphology, Viability and Germination Potential. Plant J. 2017, doi:10.1111/tpj.13667.
- Schepetilnikov, M.; Makarian, J.; Srour, O.; Geldreich, A.; Yang, Z.; Chicher, J.; Hammann, P.; Ryabova, L.A. GTP Ase ROP 2 Binds and Promotes Activation of Target of Rapamycin, TOR, in Response to Auxin. EMBO J. 2017, doi:10.15252/embj.201694816.
- Jones, M.A.; Shen, J.J.; Fu, Y.; Li, H.; Yang, Z.; Grierson, C.S. The Arabidopsis Rop2 GTPase Is a Positive Regulator of Both Root Hair Initiation and Tip Growth. Plant Cell 2002, doi:10.1105/tpc.010359.