
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Pavel Hozák | + 2429 word(s) | 2429 | 2021-07-05 11:04:33 |
Video Upload Options
Super-resolution microscopy (SRM) achieves sub-diffraction limited resolution by either deterministic or stochastic time-controlled emission of fluorescence from only a subset of fluorophores from the total fluorophore population in the specimen. Single-molecule localization microscopy (SMLM) allows for the visualization of individual fluorophores at the level of individual molecules based on the temporal separation of individual fluorescent molecules. This relatively simple principle allows for the detection at subsequent time intervals of a sparse subset of well-separated fluorophores and reconstruction of a final super-resolved image from the localization of the centers of the fluorophores.
1. Introduction
The optical resolution of the conventional light microscopy is limited by the diffraction of light and allows to distinguish the objects only if they are ~200 nm apart. This ~200 nm resolution limit of the classical light microscope is given by the nature of light and results from the fundamental laws of physics. Ernst Abbe postulated in 1873 that the limit of discrimination will never pass significantly beyond half the wavelength of blue light [1][2]. Reducing the wavelength of the light used for imaging and/or increasing the numerical aperture (NA) of the objective lens improves the spatial resolution of a conventional light microscope. Lord Rayleigh postulated in 1896 that the smallest resolvable distance between two points is proportional to the wavelength in the vacuum of the light used for imaging and inversely proportional to the NA with a factor of 0.61 [3]. Hence, it is possible to improve the resolution by shortening the wavelength, and therefore, use of electrons instead of light allowed for the visualization of the biological specimens at the ultrastructural level.
Super-resolution microscopy (SRM), contrary to electron microscopy, allows for live-cell imaging at the previously unprecedented spatial and temporal level. Novel and progressive SRM approaches helped to uncover many previously undetected dynamic mechanisms in living cells and provided valuable contextual information about cellular nanostructures.
2. SRM Approaches at a Glance
SRM achieves sub-diffraction limited resolution by time-controlled emission of fluorescence from only a subset of fluorophores from the total fluorophore population in the specimen. Two main concepts of SRM are (i) reversible saturable optical fluorescence transitions (RESOLFT) that allow deterministic temporal control of fluorophore emission, and (ii) single-molecule localization microscopy (SMLM) based on the stochastic temporal control of fluorophore emission [4][5].
2.1. Deterministic SRM Approaches: SIM and STED
The RESOLFT family [6][7][8][9] includes saturated structured illumination microscopy (S-SIM) [10], which is an upgrade of the previously structured illumination microscopy (SIM) [11]. In the classical SIM, Gustafsson implemented high-frequency line-patterned illumination and improved axial and lateral resolution by a factor of two [11][12]. The subsequent 3D SIM improved spatial resolution by an additional factor of two [13][14][15]. However, classical SIM did not demonstrate the potential to increase the resolution without limit, and therefore the classification of SIM as a typical SRM varies [16][4][5]. Nevertheless, S-SIM, which is a nonlinear modification of classical SIM, is in theory, capable of unlimited resolution [10] and is also applicable for live-cell imaging [17]. A typical representative of the RESOLFT family is the stimulated emission depletion (STED) microscopy and its variations [18]. The resolution of a confocal microscope, on which STED is typically based, is increased by attenuating the fluorescence at the periphery of the excitation spot. The fluorescently labeled sample is illuminated by an excitation laser simultaneously with the depletion laser that has a wavelength within the emission spectrum of the imaged fluorophore. The depletion laser beam has a ring or donut shape and is superimposed over the excitation laser beam. The depletion laser at a certain intensity, called saturation intensity, then switches off the fluorescence over a controlled area at the periphery of the excitation spot by a process called stimulated emission [19] and effectively reduces the diameter of a recorded fluorescence spot.
2.2. Stochastic SRM Approaches: Single-Molecule Localization Microscopy
SMLM, in contrast to the above-discussed deterministic SRM techniques, allows for the visualization of individual fluorophores at the level of individual molecules. This revolutionary approach is based on the temporal separation of individual fluorescent molecules based on a relatively simple principle that allows for the detection at subsequent time intervals of a sparse subset of well-separated fluorophores and reconstruction of a final super-resolved image from the localization of the centers of the fluorophores (Figure 1).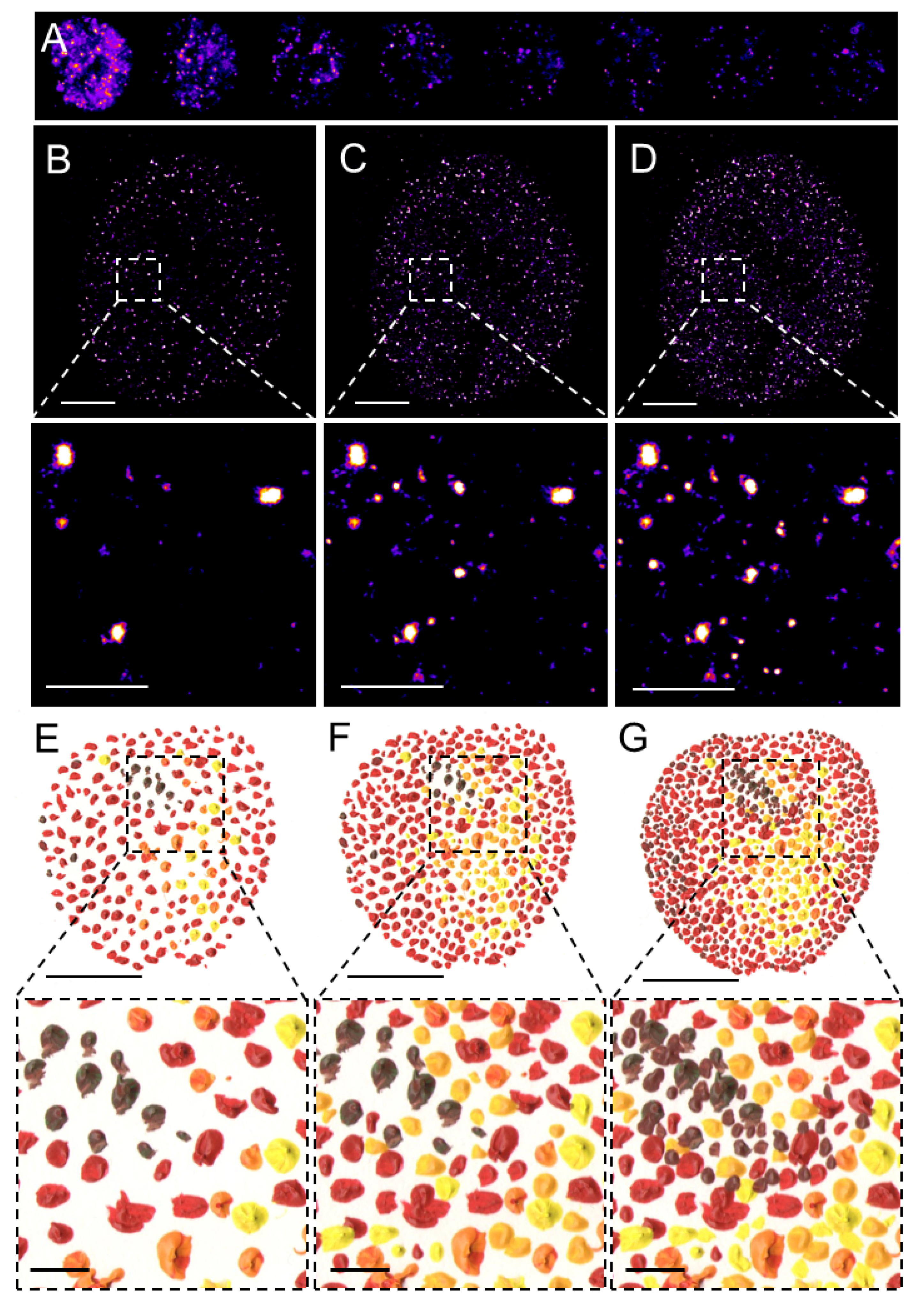
Figure 1. Pointillistic nature of single-molecule localization microscopy. (A) Individual frames from the time-lapse acquisition of raw blinking of Alexa Fluor (AF)647-labeled secondary antibody against primary anti-RNAPII antibody (RNAPII-AF647). The signal of individual AF647 molecules accumulates in time (B,C) to create the final dSTORM image of RNAPII (C). The imaging process in (B–D) resembles an artistic painting style called pointillism in which the accumulation of color spots on a canvas (E,F) creates the final painting (G). Artistic paintings in (E–G) were kindly provided by Katarína Mrvová. White bar = 5 µm; inset 1 µm. Black bar = 5 cm; inset 1 cm.
SMLM techniques originally included photoactivated or fluorescence photoactivation localization microscopy (PALM) [20] or FPALM [21] and stochastic optical reconstruction microscopy (STORM) [22] or direct STORM (dSTORM) [23][24][25] and their modifications. These methods limit the number of fluorophores that emit photons at the same time and assemble the final super-resolved image from sequentially emitting fluorophores. Reduced probability of simultaneous detection of overlapping signals in one acquisition frame allows localizing the center of individual emitters with the precision in the range of lower tens of nanometers, depending on the number of emitted photons. PALM utilizes the photo-activatable fluorescent proteins (PA-FPs) that can be reversibly switched between the non-fluorescent and fluorescent states or the photo-convertible FPs (PC-FPs) that can be switched between two fluorescent states with different excitation and emission pattern [26][27]. Individual molecules of the PA- and PC-FPs are stochastically photo-activated and photo-converted, respectively, while neighboring molecules remain non-fluorescent or undetected and therefore individual FP molecules can be sequentially localized at different time intervals. Fluorescence emitted by the individual and spatio-temporaly well separated PA or PC form of an FP is detected by sensitive cameras and fitting of the Gaussian profile to the signal enables to measure the position of a molecule with a precision far better than in the diffraction-limited conventional fluorescence microscopy. The principle of reversible stochastic photo-switching of organic fluorochromes between the fluorescent and long-lived dark states was first utilized in STORM using photo-switchable dye pairs [22]. This principle allowed for the detection of a sparse subset of fluorochromes in their fluorescent “on” state that is well separated at a given time-point. Thus, the precise localization, e.g., the center of the mass, of individual particles can be determined with high precision and this process is iteratively repeated (Figure 1A). The final super-resolved image is created by the sum of individual single-molecule localization (Figure 1B–D) and therefore resembles of artistic painting technique pointillism (Figure 1E–G) [28]. Soon after its introduction, STORM was implemented for 3D [29][30] and live-cell imaging [31]. In contrast to PALM and STORM that use PA-FPs and a combination of activator and reporter pairs of organic fluorochromes, respectively, dSTORM uses conventional fluorescent probes such as labeled antibodies or self-labeling tags [23][25].
3. Progressive Development of Fluorophores for SMLM
The rapid development of live-cell SMLM applications is linked to the progressive development of fluorophores with specific and unprecedented photophysical properties. Fluorophores used in SMLM can either be genetically encoded FPs, “self-labeling” tags such as HaloTag or SNAP that covalently associate with organic fluorochromes or organic fluorochromes such as Alexa Fluores attached directly to a primary antibody or attached to a secondary antibody directed against the primary antibody as in classical immunofluorescence protocols (Figures 1). A plethora of conventional fluorophores, such as Alexa Fluor 647 that is the most commonly used in dSTORM [31][32], are suitable for SMLM [33]. The advantage of rhodamine-based fluorophores is that they exist in a dynamic equilibrium between a fluorescent zwitterion and a non-fluorescent but cell-permeable spirocyclic form. This feature of rhodamine dyes and their derivatives can be differently utilized in various SRM modalities that have different requirements of the dynamic equilibrium between the zwitterion and spirocyclic rhodamine forms [34][35].
As mentioned above, individual fluorescent proteins are randomly photo-activated or photo-converted at different time points in PALM, while their neighbors remain dark or undetected [20][21][26]. The monomeric members of the Eos family of the PC-FPs [34] provide high brightness with good contrast and are suitable for live-cell SRM [20][26]. Photo-convertible fluorescent protein mEos2 is an improvement of its parent protein mEosFP [36] that was not suitable for use in mammalian cells due to its maturation at lower temperatures. Utilization of mEos2 facilitated, for instance, the single-molecule kinetics study of transcription factors (TFs) glucocorticoid receptor (GR) and estrogen receptor-α (ER) [37]. Photo-convertible fluorescent protein Dendra2 [38] is initially green-emitting and upon 405 nm illumination, converts into a red-emitting form [39][40]. Illumination with the very low (~1 W/cm2) intensity of 405 nm light allows for the photo-conversion of only a small subset of Dendra2 molecules in the sample. Favorable photo-physics [41] and low aggregation propensity [42] of Dendra2 made it optimal to study, for instance, the dynamics of RPB1, the large catalytic subunit of RNAPII [43][44][45][46], transcription coactivator Mediator [46] or TFs c-Myc and P-TEFb [47]. However, the photo-converted Dendra2 molecules remain in the red-emitting state for several tens or even hundreds of milliseconds, and therefore, a single molecule may appear in multiple acquisition frames. Moreover, in addition to irreversible photo-bleaching, Dendra2 molecules undergo intermittent photo-physical blinking transitions [41][48] that obscure direct correlation between counts of detections and exact numbers of molecules [44].
HaloTag and SNAP tagged proteins exhibited stable binding events in the nucleus [47] but these unspecific binding events were separated from the specific binding events of the proteins of interest [49]. A comparative SRM study of tagged TF favored HaloTag in terms of unspecific binding, the photostability of the conjugated fluorophore, or localization precision [50]. The discovery of a fine-tuning method for the development of new and highly photostable fluorophores called Janelia Fluors (JFs) [51] for live-cell SRM revolutionized the field and opened almost unlimited opportunities to explore living systems at high spatial and temporal resolution. JFs are available as reactive organic molecules for direct labeling or as HaloTag and SNAP substrates. An important advantage of many of the JFs is their fluorogenicity, e.g., a significant increase in fluorescence upon their binding to HaloTag or SNAP, which alleviates the necessity of extensively washing out unbound fluorophores and reduces unspecific background [52]. Some of the most frequently used JFs so far were JF549, which is a prototypical JF and a direct analog of tetramethyl rhodamine (TMR) or its silicone-containing far-red counterpart JF646 [51][53]. Photoactivatable versions of JF549 and JF646 allow to switch on only a subset of fluorophores, which is advantageous for single-particle tracking (SPT) in living cells [54].
The development of PA fluorochromes extended the possibilities previously offered by PA- or PC-FPs. However, the dependence on the photoactivation for SMLM and SPT can be genetically circumvented by the precise copy number control of the fluorescently labeled fusion proteins [55]. This approach is particularly useful for the imaging of individual molecules in the densely packed nucleus. Progressive development of finely-tuned fluorophores [35] including fluorophores with very specific features, such as photosensitizers that generate reactive oxygen species [56] for delicate applications and allowing for multiplexing at a high spatial and temporal resolution [34] continues to push the boundaries of SMLM applications. Moreover, the fluorescent signal of individual molecules of interest can be amplified by multivalent tags such as Sun-tag [57] or repetitive HaloTag [58][55].
3. Dynamics of RNAPII by SMLM
Transcription by RNAPII is essential for gene expression and therefore fundamental for cellular activities. RPB1 enzyme is the large catalytic subunit of RNAPII central to the transcription process. In humans, RPB1 C-terminal domain (CTD) contains 52 heptad repeats that can be reversibly phosphorylated and the specific phosphorylation pattern reflects subsequent steps of transcription [59][60][61]. RPB1 forms clusters with a half-life in the order of seconds [43], which is significantly faster than the several minutes required to complete the transcription of a typical mammalian gene. Upon the CTD phosphorylation RPB1 switches into the elongation phase, RPB1 clusters dissolve, indicating that elongating RNAPII leaves clusters formed at the (pre-) initiation phase [62]. Therefore, the dynamic clustering of RPB1 is linked to the assembly of macromolecular complexes during pre-initiation and initiation steps of transcription. Short-lived clusters observed on the actively transcribed gene are transient agglomerations of ~80 polymerases, which are distinct from the much lower counts of elongating polymerases that likely appeared as low frequency of detections [44].
In summary, several reviewed SMLM approaches enabled us to study the RNAPII transcription in unprecedented spatial and temporal details and thereby helped to finely refine our mechanistic understanding of the regulation of gene expression. The dynamic clustering of RPB1 before productive transcription elongation and the assembly of macromolecular complexes during pre-initiation and initiation steps of transcription revealed by SMLM has implications for the fine regulation of gene expression. Integration of the information obtained by the SMLM together with biochemistry helped to build a mechanistic model of the localized hierarchical recruitment of co-activators and TFs during the pre-initiation phase followed by the recruitment and formation of RNAPII clusters during the initiation phase and followed by RNAPII leaving the clusters during elongation phase. This hierarchical and dynamic principle of the transcriptional regulation and the discovery of RNAPII forming transient condensates that dictates the productive transcriptional output has progressively changed our mechanistic understanding of gene expression.
4. Conclusions and Perspectives
Since the publication of one of the seminal reviews on SRM in 2010 [4], twelve years have passed during which the expectation of authors that “it will still take time and further engineering until (the) technical developments (of SMLM) find their way into commercial systems” has become a reality and in the past decade we have witnessed rapid progress in the 4D single-molecule kinetic studies in living cells. Nevertheless, many secrets of the establishment and propagation of the living matter in space and time remain unexplored. The progressive development of SRM, visualization tools and SMLM in particular will continue to help cell biologists to uncover these secrets. Moreover, the combination of SMLM with other imaging approaches such as EM will continue to provide valuable contextual information about cellular nanostructures [20][63]. The relatively new but rapidly growing field of nuclear lipid biology will greatly benefit from the progressive development of quantitative single-molecule imaging approaches [64][65][66][67][68][69][70][71][72][73][74]. Although a variety of tools is applicable for the specific visualization of various lipid species in the membranes, lipids in the membranes display 2D distributions and dynamics that differ from the nuclear interior that is organized within membrane-less sub-compartments [75]. Therefore, the development of the tools for visualization of specific nuclear lipids by single-molecule imaging together with the use of novel fluorophores and implementation of the 3D SPT will be crucial for the kinetic studies of the nuclear lipids and their roles in the establishment of the functional nuclear architecture, regulation of transcription and overall gene expression.




