Estrogen nuclear receptors, represented by the canonical forms ERα66 and ERβ1, are the main mediators of the estrogen-dependent pathophysiology in mammals. However, numerous isoforms have been identified, stimulating unconventional estrogen response pathways leading to complex cellular and tissue responses. The estrogen receptor variant, ERα36, was cloned in 2005 and is mainly described in the literature to be involved in the progression of mammary tumors and in the acquired resistance to anti-estrogen drugs, such as tamoxifen. Nevertheless, recent data specify the place that ERα36 currently occupies within the diversity of nuclear and membrane estrogen receptors and highlight the impact of ERα36 expression and/or activity in normal breast and testicular cells, but also in different types of tumors including mammary tumors. Therefore, ERα36 can now be considered as a marker of malignancy and the regulation of ERα36 expression could provide new clues to counteract resistance to cancer treatments in hormone-sensitive tumors.
The promoter region of the
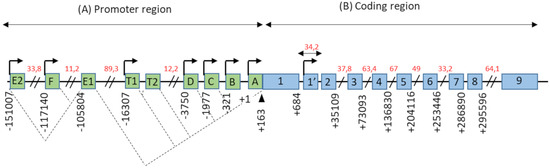
ESR1 gene spans more than 150 kb on chromosome 6 (6q25.1), and contains at least nine non-coding exons and seven alternative promoters which contribute to regulating the differential expression of ERα in different estrogen target tissues (). The first transcription start site (TSS), located at the level of the non-coding exon “A”, was identified by Greene and colleagues in 1986 [
2]. In addition, Keaveney and colleagues [
13] demonstrated the existence of an acceptor splice site localized 163 bp downstream of the exon A TSS. This site is highly conserved among vertebrates as the point of anchorage for the various non-coding exons of the promoter region. Then, the majority of non-coding exons were shown to have their own TSS [
14]. Moreover, an eighth alternative promoter located upstream of exon 1 has been described [
14,
15,
16].
Figure 1. Genomic organization of the
ESR1 gene. (
A) The promoter region of the
ESR1 gene contains 9 non-coding exons (green boxes), annotated from A to F—T1, T2, and 7 alternative promoters each having its own TSS (arrows). There is also a splice acceptor site located at position +163 bp. The dotted lines symbolize the different possibilities for the splicing of non-coding exons, as described in the literature. Exons E1 and T2 expression depends on the promoters E2 and T1, respectively. (
B) The coding region of the
ESR1 gene consists of 10 exons (blue boxes), numbered from 1–1′to 9, separated by 9 introns. There is also an 8th alternative promoter located at the level of exon 1′. In this schematic representation, the scale is not respected but some estimated positions are indicated below the exons, in base pairs (bp). Position +1 corresponds to the first transcription initiation site (TSS) described by Greene and colleagues, in 1986 [
2]. The indications in red correspond to the length of long intronic regions in kilo base pairs (kbp). Adapted from [
14,
15].
The coding region of the
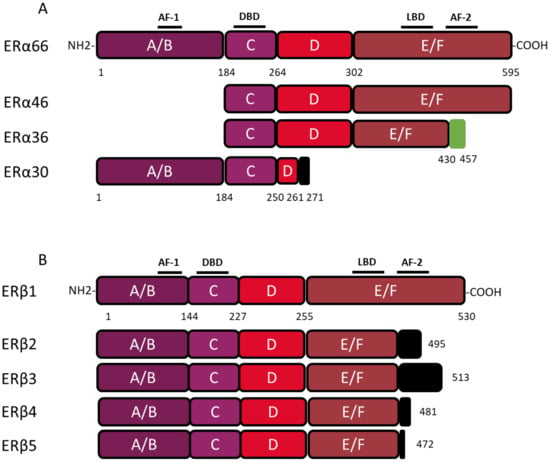
ESR1 gene spans over 300 kb and consists of 10 exons and nine introns, which are the source of four isoforms: ERα66, ERα46, ERα36, and ERα30. The canonical form of ERα, called ERα66, is a 595 amino acid protein of 66 kDa. ERα66 displays all of the functional domains typical for nuclear receptors [
17]. The A/B domain located at the N-terminal end is the least conserved domain between the members of the superfamily of nuclear receptors, both in terms of length and of amino acid sequence homology. This region contains the AF-1 transactivation domain that allows ligand-independent activation of transcription. In addition, the A/B domain has several phosphorylation and sumoylation sites that allow the recruitment of co-activators or co-repressors and can modulate the regulation of ERα66 target genes [
18]. The C domain, also called DBD (DNA-binding domain), is a highly conserved domain which allows the attachment of ERα66 to specific response elements located in the promoter region of the target genes. This domain consists of 2 zinc finger motifs, each consisting of four cysteines surrounding a Zn
2+ ion. At the base of the first zinc finger is a P-box (proximal box) motif involved in the recognition of the response elements, while the second zinc finger is associated with a D-box (distal box), which is necessary for ERα66 dimerization and function [
19,
20,
21]. The D domain is a flexible hinge region that allows the rotation of the DBD to facilitate the binding of the receptor to its response element. The nuclear localization signal that allows the translocation of the receptor to the nucleus is present in this poorly conserved region. This sequence is exposed as a result of a change in the D domain conformation induced by ligand binding [
22]. The E/F domain is the ligand-binding domain (LBD) and participates in the dimerization of the receptor. For most nuclear receptors, including ERα66, this E/F domain consists of 12 α helices and an antiparallel beta sheet, which form the binding pocket for hydrophobic ligands. This region also contains the AF-2 transactivation domain, which consists of the H3, H4, and H12 helices, and which results in the ligand-dependent activation of transcription. Indeed, depending on the agonist or antagonist nature of the ligand, the orientation of the H12 helix is different, and has an impact on the recruitment of the co-factors needed to regulate ERα66 transcriptional activity [
23,
24].
The three variants of ERα66, namely ERα46, ERα36, and ERα30, result from alternative splicing mechanisms, but also from the use of alternative promoters (). Unlike the canonical form ERα66, which is encoded by exons 1–8, the variant ERα46 is transcribed from promoters E and F and encoded from exons 2 to 8. At the protein level (), this 46 kDa variant lacks the AF-1 transactivation domain. ERα46 can be palmitoylated by the PAT (palmitoyltransferase) enzyme and thus recruited to the caveolae at the plasma membrane [
25]. This variant triggers more potent non-genomic signaling than ERα66 [
10]. Moreover, ERα46 expression in ERα-positive tumors is associated with less aggressive tumors, likely due to the fact that it decreases the proliferation rate in response to E
2 [
26].
More recently, a 30 kDa variant, called ERα30, was identified by Zhu H. and colleagues [
27]. This variant is encoded by exons 1, 2, 3, and 8, but also by a part of exons 4 and 6. In effect, during alternative splicing, the first 24 nucleotides of exon 4 are directly linked to the last 44 nucleotides of exon 6. At the protein level, it results in a truncated D hinge domain as well as in the absence of the LBD and the AF-2 transactivation domain. ERα30 has a small C-terminal 10 amino acid-specific region, of which the function and impact on the structure of the protein are not described [
27].
Figure 2. Structure of canonical forms and different variants of ERα66 and ERβ1. The canonical ERα 66 and ERβ1 respectively encompass 595 and 530 amino acids. (
A) The ERα66 variants, ERα46 and ERα36, do not display a A/B domain which is involved in ligand-independent transactivation. In addition, the ERα36 variant has a truncated E/F domain and a small specific sequence, of 27 amino acids (green box), at the C-terminal end. The ERα30 variant retains only the A/B domain, the C domain and part of a D domain. In the C-terminus this variant has a short specific sequence of 10 amino acids (black box). (
B) The 4 variants of ERβ, all have a truncated E/F domain and a small specific sequence of variable size at the C-terminal end (black box). Characteristic of ERβ2, the 26 amino acids specific to the C-terminal end are encoded by an exon called “exon 9” or “exon cx” located downstream of exon 8 of the
ESR2 gene. Adapted from [
28,
29].
The ERα36 variant is transcribed from an alternative promoter located in the first intron of the
ESR1 gene and is encoded by exons 1′, 2–6, and 9. From a functional point of view, the ERα36 protein, retains the DBD, as well as part of the LBD, but not AF-1 and AF-2 transactivation domains. ERα36 also possesses a specific C-terminal region of 27 amino acids encoded by exon 9 and containing a functional nuclear export signal, inducing the exportation of ERα36 from the nucleus [
15,
30]. Moreover, this unique C-terminal domain of ERα36 and more precisely the L297 residue plays a crucial role in the interaction between ERα36 and p-ERK2 [
30].