
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Joram Kiriga | + 2311 word(s) | 2311 | 2020-12-10 08:13:56 | | | |
| 2 | Peter Tang | -885 word(s) | 1426 | 2020-12-17 06:40:56 | | |
Video Upload Options
Plant non coding RNA review paper highlight the current knowledge of plant microRNAs, siRNAs, and lncRNAs, focussing on their origin, biogenesis, mode of actions, and their fundamental roles in plant response to abiotic stresses.
1. Introduction
While up to 90% of the eukaryotic genome is transcribed into RNA, only about 2% of transcribed RNAs generate protein products [1][2]. The remaining transcriptome comprises ncRNAs transcripts, which were previously considered transcriptional noise owing to the absence of protein-coding ability and poorly conserved sequences [1][3]. However, high-throughput sequencing analysis and experimental validation have confirmed the involvement of ncRNAs in a myriad of distinct gene regulatory levels, including epigenetic, transcriptional, and post-transcriptional [4].
Since their initial discovery, ncRNAs have been categorized into several distinct classes based on their origin, biogenesis, and mechanism of action. Housekeeping and regulatory ncRNAs are the major groups of ncRNA transcripts. The housekeeping ncRNAs primarily deals with cellular and ribosomal functions. They include transfer RNAs (tRNAs), small nuclear RNAs (snRNAs), ribosomal RNAs (rRNAs), and small nucleolar RNAs (snoRNAs) (Figure 1). On the other hand, the regulatory ncRNAs consist of microRNAs (miRNAs), short interfering RNAs (siRNAs), piwi-interacting RNAs (piRNAs), and long non-coding RNAs (lncRNAs) [5], (Figure 1).
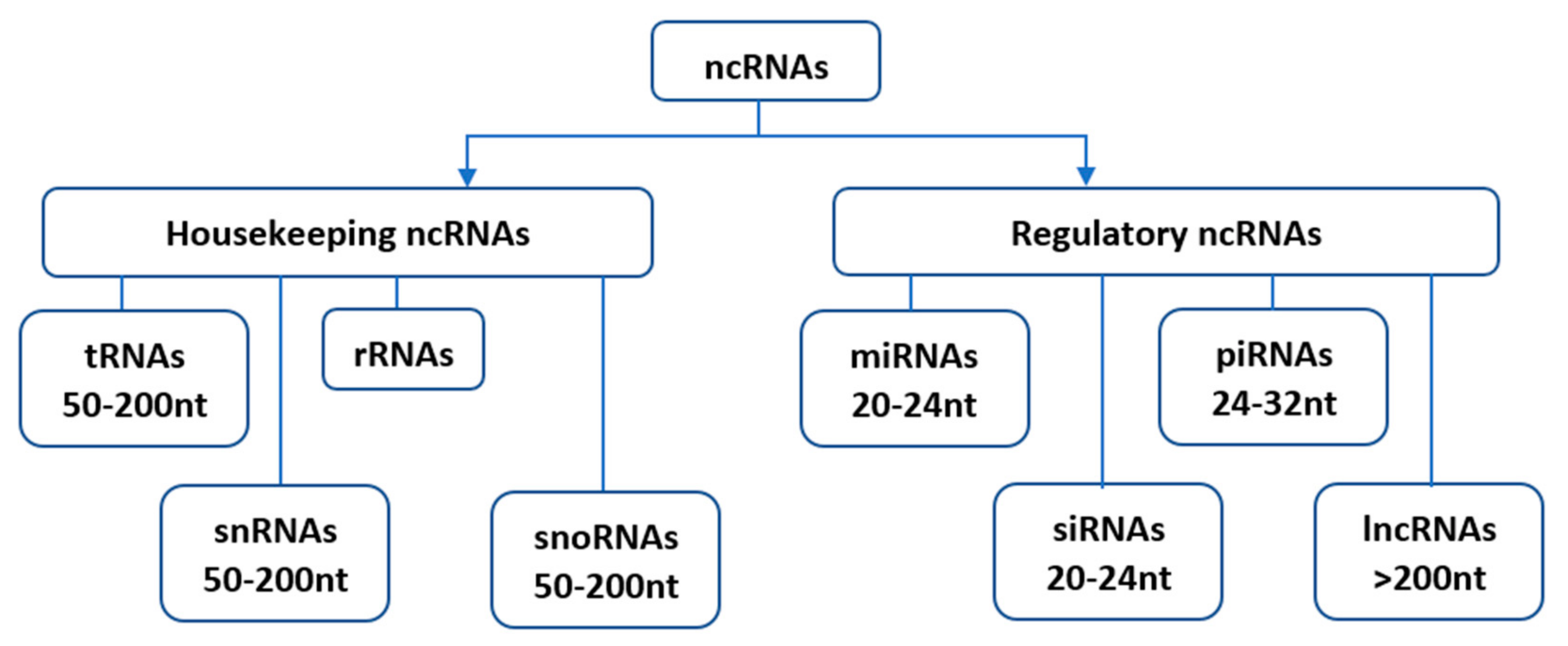
Figure 1. Classification of non-coding RNAs (ncRNAs). Housekeeping ncRNAs include; tRNAs- transfer RNAs, snRNAs-small nuclear RNAs, rRNAs-ribosomal RNAs, snoRNAs-small nucleolar RNAs. The regulatory ncRNAs consist of miRNAs-microRNAs, siRNAs-short interfering RNAs, piRNAs-piwi-interacting RNAs, and lncRNAs-long non-coding RNAs.
2. Origin and Biogenesis of Non-Coding RNAs
MicroRNAs are small but fundamental molecules which are predominantly 20–22 nt in length [6]. They are synthesized from miRNA genes (MIR genes), which are processed by RNA polymerase II (RNA pol II), RNase III Dicer-like protein 1 (DCL1) [7] (Figure 2). HASTY (HST) transports the resulting miRNA–miRNA* duplex to the cytoplasm where it is integrated into the RNA-induced silencing complex (RISC) containing ARGONAUTE 1 (AGO1) proteins that control gene silencing by mRNA cleavage and translational repression [8], (Figure 2).
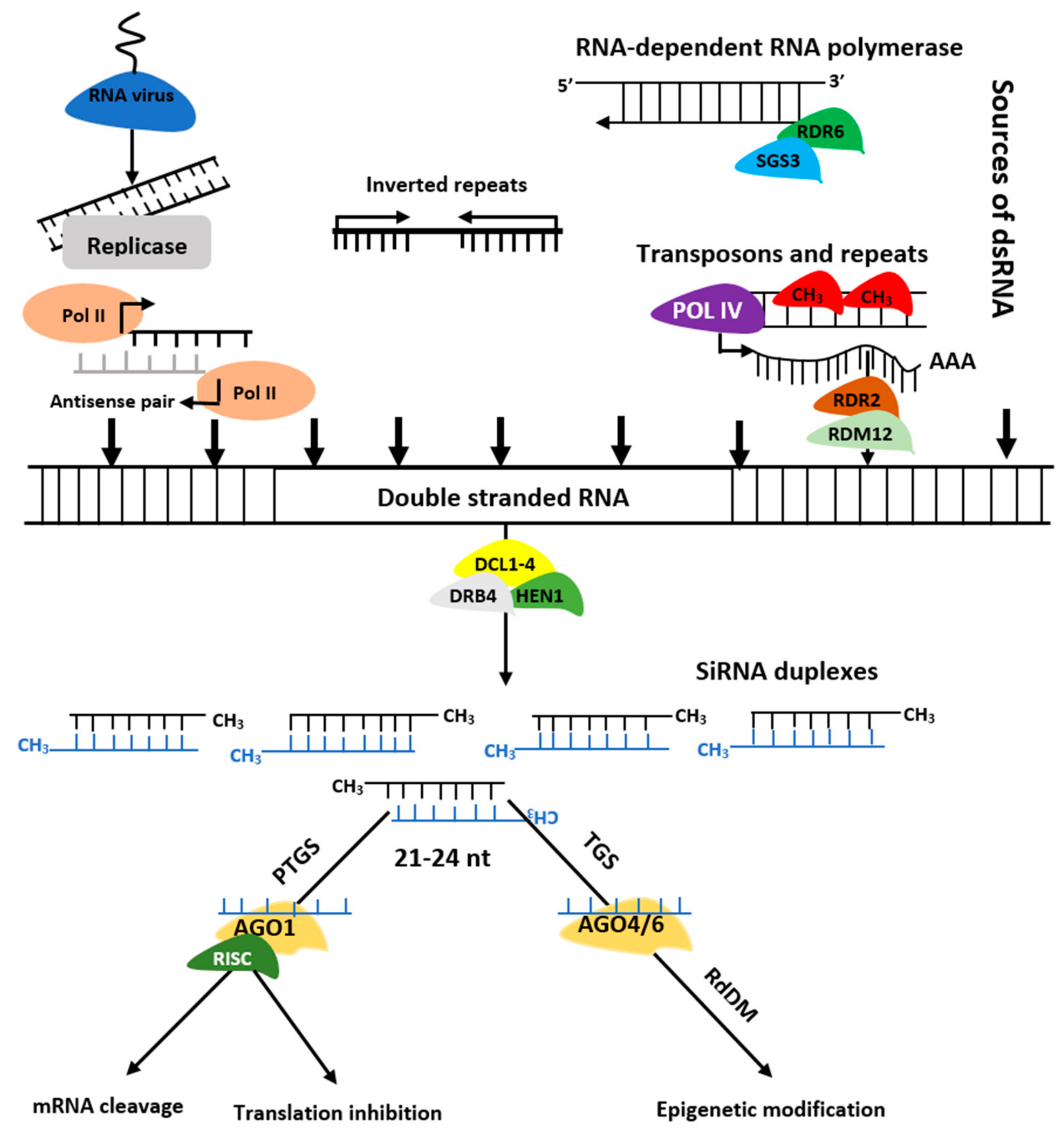
Figure 2. Biogenesis of plant miRNAs. In the nucleus, MIR genes are processed to miRNA/miRNA* duplexes through the action of Pol II, DCL1, SE, HYL1, HEN1, and HASTY. In the cytoplasm, the duplex is incorporated with RISC-AGO complex to which guides it towards the target, resulting in suppression of the translation and cleavage of mRNAs.
The siRNAs are grouped into trans-acting siRNAs (tasiRNA), heterochromatic siRNAs (hc-siRNA), and natural antisense siRNAs (nat-siRNAs) depending on the mode of action as well as biogenesis. The siRNAs biogenesis involves dsRNAs, DCL enzymes activities, and RISC containing AGO protein that controls target regulation at post-transcription or transcriptional level [9] (Figure 3). Biogenesis of ta-siRNA is dependent on SUPPRESSOR OF GENE SILENCING3 (SGS3), RNA-dependent RNA polymerases 6 (RDR6), and DCL4 [10] while that of nat-siRNAs depends on SGS3, RDR6, DCL1, DCL2, and a plant-specific RNA polymerase, NRPD1A [11]. Similarly, hc-siRNAs are generated by Pol IV, DCL3, RDR2, 5S rRNA genes, and NRPD1A [12].

Figure 3. Biogenesis of siRNAs. The double stranded RNA (dsRNAs) is transformed into siRNAs by DCL, HEN1, and DRB. The RISC-AGO complex then guides the selected strands of siRNA duplexes to post transcription gene silencing (PTGS) or transcription gene silencing (TGS).
Long non-coding RNAs are transcripts that exceed more than 200 base pair (bp) in length and cannot be translated into protein [13]. They are classified into cis-natural antisense (cis-NATs), trans-natural antisense transcripts (trans-NATs), and pseudogenes [14], sense or antisense (strand of origin) [15], divergent, or convergent (orientation of transcription), and as intronic or intergenic (location) [16] (Figure 4). They are transcribed by RNA pol II, III, IV, and V and control target regulation via multiple ways, including chromatin remodeling, transcriptional repression, mimicry, RNA splicing and transcriptional enhancer (Figure 4).
Figure 4. The biogenesis of long-non-coding RNAs (lncRNAs) and their gene regulation mechanisms in plants. (i) The transcripts of lncRNA (red box) are classified on the basis of their genomic location and in relation to the nearest gene (blue box): (A) sense lncRNAs are transcribed on the same strand as an exon; (B) antisense lncRNAs are transcribed on the opposite strand of an exon; (C) intronic lncRNAs are transcribed on the intron; (D) intergenic lncRNAs are located between two distinct genes; (E) enhancer lncRNAs emerge from an enhancer region of protein-coding genes. (ii) The gene regulation pathways induced in plants by lncRNAs. LncRNAs regulate gene expression either by: (1) interacting with transcriptional activator leading to gene activation; (2) interacting with transcriptional repressor thereby suppressing transcription; (3) controlling RNA splicing by interacting with splicing factor or binding premRNA splicing junction; (4) recruiting chromatin remodeling complex such as PRC to regulate gene expression in the promoter region; (5) LncRNA mimics miRNAs by occupying their target sites on the mRNA.
3. Impact of Non-Coding RNAs on Plant Gene Regulation
3.1. Cleavage of Target Transcripts by miRNAs and siRNAs
For miRNAs, the MIR genes are processed by DCL1, HYL1 and SE complex into mature miRNA–miRNA* duplex and transported by HASTY protein to the cytoplasm where it is integrated with RISC containing AGO1. The RISC-AGO1 complex binds to the complementary sites of sense sequence on its targeted RNA transcript and degrade it. In contrast, the antisense strand of miRNAs remains in the RISC [17]. In siRNAs, one strand which is integrated with RISC-AGO1 or AGO7 complex is guided to cleave the transcripts of the target gene at 10–11 nt upstream of the 5′ end of the antisense strand [18]. The EXONUCLEASE 4 (XRN4) enzyme then degrade both the 3′ and 5′ cleaved fragments [19]. Recent studies have reported the RNA binding and target slicer activities of AGO2, AGO4, and AGO10 in plants, indicating the complexity of the sRNA-induced gene silencing mechanism [20].
3.2. miRNAs and siRNAs Induced Translational Inhibition of Target Genes
The RISC-AGO1 complex suppresses translation by binding to the 5′ untranslated region (UTR) or open reading frame (ORF) of the target gene, thereby restricting ribosome recruitment or movement [21]. This translation inhibition process is regulated by other factors such as ALTERED MERISTEM PROGRAM 1 (AMP1), VARICOSE (VCS), GW-repeat protein, and microtubule enzyme KATANIN (KTN1) [22]. Despite the involvement of both AGO1 and AGO10, the specific function of each AGO gene in this inhibition process is still not clear [22].
3.3. miRNA and siRNA‑Directed DNA Methylation
The DCL family has multiple copies in Arabidopsis, which contribute to the biogenesis of divergent lengths of sRNA. DCL1 is involved in the conversion of partially paired dsRNAs precursors into 21-nt mature miRNAs [23]. On the other hand, DCL2 and DCL44 initiate the generation of 20–22nt siRNAs from perfect complementary dsRNAs precursors [24]. DCL3 produces 24-nt siRNAs (hc-siRNA) that typically silence gene expression via the RdDM pathway [25]. These hc-siRNAs are transcribed at the heterochromatic regions where they stimulate cytosine methylation in the sequence contexts of CG, CHG, and CHH, in cis [26]. The DCL3-dependent miRNAs integrate with AGO4 forming a complex which repress gene silencing by cytosine and histone methylation [27]. However, the de novo hc-siRNA-induced RdDM process integrates the activities of RDR, DCL, AGO, and Pol IV and V which transcribe the double-stranded precursors and promote methylation at the target sites respectively. Therefore, DNA and lysine methylation at the ninth position of histone H3 (H3K9) induces systemic silencing, while the hc-siRNA/AGO4 RISC guides DNA and H3K9 methyltransferases to the target sequence for transcriptional gene silencing [26].
3.4. Gene Expression Regulation by lncRNAs
Plant lncRNAs function in cis and trans. The cis-acting lncRNAs function near their synthesis sites and operate directly on the local nucleotide sequences or chromosome regions on one or more contiguous genes. The trans-acting lncRNAs, on the other hand, disperse from the synthesis site and can function on several genes at great distances, including on different chromosomes [28]. Moreover, lncRNAs can also be transcribed as sRNAs precursors [29], miRNA target mimics where the lncRNAs competes with the target mRNA for miRNA binding [30], protein-protein interactions, modification, localization [31], and via epigenetic regulation mechanisms such as DNA methylation, histone modification, and chromatin remodeling [32].
4. Function of ncRNAs in Response to Abiotic Stress in Plants
Plants have developed various mechanisms to adjust their growth and development during environmental stress conditions. Multiple research findings have recently shown the differential expression of ncRNAs in such unfavourable conditions [33]. These ncRNAs either control gene expression in related cellular networks or function directly in response to stress [34]. miRNAs have become new targets for improving plant productivity and abiotic stress tolerance due to the broad functions of their targets [35]. The regulation of miRNAs appears to rely on their roles on the abiotic stress response. Stress-up-regulated miRNAs might down-regulate their target genes, which might be negative regulators of stress tolerance (e.g. stress-responsive gene repressors) [36]. Moreover, the down-regulation of miRNAs under stress might accumulate their target gene mRNAs, which might positively regulate stress tolerance [36]. Moreover, various siRNAs and lncRNAs have been reported in response to multiple abiotic stress [33]. All these ncRNAs have been reported to execute their functions in responding to abiotic stresses in different ways.
References
- Pauli, A.; Rinn, J.L.; Schier, A.F. Non-coding RNAs as regulators of embryogenesis. Nat. Rev. Genet. 2011, 12, 136–149.
- Rai, M.I.; Alam, M.; Lightfoot, D.A.; Gurha, P.; Afzal, A.J. Classification and experimental identification of plant long non-coding RNAs. Genomics 2019, 111, 997–1005.
- Ariel, F.; Romero-Barrios, N.; Jégu, T.; Benhamed, M.; Crespi, M. Battles and hijacks: Noncoding transcription in plants. Trends Plant Sci. 2015, 20, 362–371.
- Cech, T.R.; Steitz, J.A. The noncoding RNA revolution-trashing old rules to forge new ones. Cell 2014, 157, 77–94.
- Morris, K.V.; Mattick, J.S. The rise of regulatory RNA. Nat. Rev. Genet. 2014, 15, 423–437.
- Jones-Rhoades, M.W.; Bartel, D.P. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell 2004, 14, 787–799.
- Axtell, M.J.; Meyers, B.C. Revisiting Criteria for Plant MicroRNA Annotation in the Era of Big Data. Plant Cell 2018, 30, 272–284.
- Pareek, M.; Yogindran, S.; Mukherjee, S.K.; Rajam, M.V. Plant MicroRNAs: Biogenesis, Functions, and Applications. In Plant Biology and Biotechnology; Springer: New Delhi, India, 2015, 639–661.
- Erdmann, R.M.; Picard, C.L. RNA-directed DNA Methylation. PLoS Genet. 2020, 16, e1009034.
- Gasciolli, V.; Mallory, A.C.; Bartel, D.P.; Vaucheret, H. Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr. Biol. 2005, 15, 1494–1500.
- Borsani, O.; Zhu, J.; Verslues, P.E.; Sunkar, R.; Zhu, J.K. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 2005, 123, 1279–1291.
- Mallory, A.C.; Vaucheret, H. Functions of microRNAs and related small RNAs in plants. Nat. Genet. 2006, 38, S31–S36.
- Kapranov, P.; Cheng, J.; Dike, S.; Nix, D.A.; Duttagupta, R.; Willingham, A.T.; Bell, I. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 2007, 316, 1484–1488.
- Nie, L.; Wu, H.-J.; Hsu, J.-M.; Chang, S.-S.; Labaff, A.M.; Li, C.-W.; Wang, Y.; Hsu, J.L.; Hung, M.-C. Long non-coding RNAs: Versatile master regulators of gene expression and crucial players in cancer. Am. J. Transl. Res. 2012, 4, 127–150.
- Nam, J.W.; Bartel, D.P. Long noncoding RNAs in C. elegans. Genome Res. 2012, 22, 2529–2540.
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789.
- Lelandais-Brière, C.; Sorin, C.; Declerck, M.; Benslimane, A.; Crespi, M.; Hartmann, C. Small RNA diversity in plants and its impact in development. Curr. Genom. 2010, 11, 14–23.
- Liu, T.; Zhang, L.; Chen, G.; Shi, T. Identifying and Characterizing the Circular RNAs during the Lifespan of Arabidopsis Leaves. Front. Plant Sci. 2017, 8, 1278.
- Ren, G.; Xie, M.; Zhang, S.; Vinovskis, C.; Chen, X.; Yu, B. Methylation protects microRNAs from an AGO1-associated activity that uridylates 5′ RNA fragments generated by AGO1 cleavage. Proc. Natl. Acad. Sci. USA 2014, 111, 6365–6370.
- Zhu, H.; Hu, F.; Wang, R.; Zhou, X.; Sze, S.-H.; Liou, L.W.; Barefoot, A.; Dickman, M.; Zhang, X. Arabidopsis Argonaute10 specifically sequesters miR166/165 to regulate shoot apical meristem development. Cell 2011, 145, 242–256.
- Iwakawa, H.O.; Tomari, Y. Molecular insights into microRNA mediated translational repression in plants. Mol. Cell 2013, 52, 591–601.
- Li, S.; Liu, L.; Zhuang, X.; Yu, Y.; Liu, X.; Cui, X.; Ji, L.; Pan, Z.; Cao, X.; Mo, B.; et al. MicroRNAs inhibit the translation of target mRNAs on the endoplasmic reticulum in Arabidopsis. Cell 2013, 153, 562–574.
- Denli, A.M.; Hannon, G.J. RNAi: An ever-growing puzzle. Trends Biochem. Sci. 2003, 28, 196–201.
- Bouché, N.; Lauressergues, D.; Gasciolli, V.; Vaucheret, H. An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J. 2006, 25, 3347–3356.
- Creasey, K.M.; Zhai, J.; Borges, F.; Van Ex, F.; Regulski, M.; Meyers, B.C.; Martienssen, R.A. miRNAs trigger widespread epigenetically activated siRNAs from transposons in Arabidopsis. Nature 2014, 508, 411–415.
- Xie, Z.; Johansen, L.K.; Gustafson, A.M.; Kasschau, K.D.; Lellis, A.D.; Zilberman, D.; Jacobsen, S.E.; Carrington, J.C. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004, 2, E104.
- Ye, R.; Wang, W.; Iki, T.; Liu, C.; Wu, Y.; Ishikawa, M.; Qi, Y. Cytoplasmic assembly and selective nuclear import of Arabidopsis Argonaute4/siRNA complexes. Mol. Cell 2012, 46, 859–870.
- Lee, J.T. Epigenetic regulation by long noncoding RNAs. Science 2012, 338, 1435–1439.
- Lauressergues, D.; Couzigou, J.M.; San Clemente, H.; Martinez, Y.; Dunand, C.; Bécard, G.; Combier, J.P. Primary transcripts of microRNAs encode regulatory peptides. Nature 2015, 520, 90–93.
- Liu, J.; Wang, H.; Chua, N.H. Long non coding RNA transcriptome of plants. Plant Biotechnol. J. 2015, 13, 319–328.
- Audas, T.E.; Jacob, M.D.; Lee, S. Immobilization of proteins in the nucleolus by ribosomal intergenic spacer noncoding RNA. Mol. Cell 2012, 45, 147–157.
- Matzke, M.A.; Mosher, R.A. RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nature Reviews Genetics 2014, 15, 394-408.
- Jha, U.C.; Nayyar, H.; Jha, R.; Khurshid, M.; Zhou, M.; Mantri, N.; Siddique, K.H. Long non-coding RNAs: Emerging players regulating plant abiotic stress response and adaptation. BMC Plant Biol. 2020, 20, 1–20.
- Helliwell, C.A.; Robertson, M.; Finnegan, E.J.; Buzas, D.M.; Dennis, E.S. Vernalization-repression of Arabidopsis FLC requires promoter sequences but not antisense transcripts. PLoS ONE 2011, 6, e2151.
- Pandita, D. Plant MIRnome: miRNA Biogenesis and Abiotic Stress Response. In Plant Abiotic Stress Response; Springer: Berlin/Heidelberg, Germany, 2019, 449–474.
- Sunkar, R.; Chinnusamy, V.; Zhu, J.; Zhu, J.K. Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci. 2007, 12, 301–309.





