
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Ray Rose | + 2018 word(s) | 2018 | 2021-05-26 11:23:12 | | | |
| 2 | Catherine Yang | Meta information modification | 2018 | 2021-05-31 05:13:05 | | |
Video Upload Options
Plant mitochondria have large genomes to house a small number of key genes. Most mitochondria do not contain a whole genome. To maintain the mitochondrial genes, so important for energy production, the fusion and fission of mitochondria is critical. The dynamin related proteins DRP 3A and 3B drive the fission process. Fusion is less well understood but the MIRO2 gene appears to have a significant role. Massive mitochondrial fusion and subsequent fission prior to flowering, in the enlarging zygote and at germination aids genome repair, conservation of critical genes and may give an energy boost to key stages of the life cycle.
1. Introduction
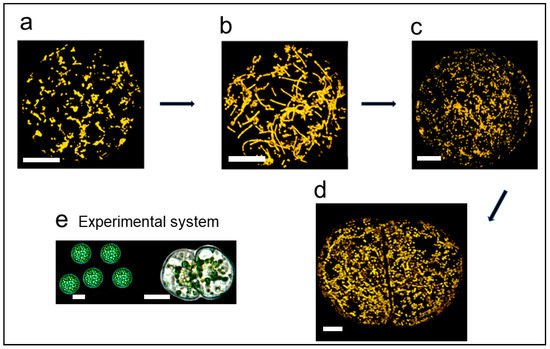
2. Mitochondrial Fusion
2.1. Demonstration of Mitochondrial Fusion
2.2. The Mechanism of Mitochondrial Fusion
3. Significance of the Mitochondrial Fusion/Fission Cycle
3.1. Mitochondrial DNA Content per Mitochondrion Is Highly Variable
3.2. MtDNA Recombination
3.3. Cytoplasmic Male Sterility
3.4. Mitochondrial Fusion and Energetics
3.5. Mitochondrial Fusion and Evolution
4. Massive Mitochondrial Fusion (MMF)
Given that the total mtDNA of the cell must be considered as a single entity, this makes MMF or hyperfusion an important part of maintaining the integrity of the mitochondrial genome. This means that it provides an important opportunity for all the subgenomes to interact for recombination and DNA repair for the next generation. What is known currently is that MMF occurs in the SAM where flowering is initiated, in the zygote and in germination, which are key points in the life cycle. This is not to say that the fusion/fission cycle involving few mitochondria is not unimportant in the cell cycle, cell development and the functioning of the cell. In these latter cases, the importance may be in DNA replication, and ensuring transcripts, proteins and metabolites are readily available for the maintenance of functional mitochondria and their genomes.
While the fusion/fission cycle is of key importance for maintaining mitochondria and their genome, there may be other roles for MMF. In plants, there is some evidence that fusion favours high energy demand and MMF occurs at times prior to the onset of major development shifts. MMF also occurs prior to the first cell division on the path to regeneration. If there is a connection between MMF and ATP production, there may be a role for manipulating mitochondrial fusion as an approach to modulating mitochondrial performance. In mammalian cells there is evidence that the hyperfused mitochondrial reticulum in the GI/S stage of the cell cycle produces more ATP than any other stage of the cell cycle.
References
- Rose, R.J. Sustaining life: Maintaining chloroplasts and mitochondria and their genomes in plants. Yale J. Biol. Med. 2019, 92, 499–510.
- Mackenzie, S.; McIntosh, L. Higher plant mitochondria. Plant Cell 1999, 11, 571–585.
- Smith, A.G.; Croft, M.T.; Moulin, M.; Webb, M.E. Plants need their vitamins too. Curr. Opin. Plant Biol. 2007, 10, 266–275.
- Sweetlove, L.J.; Fait, A.; Nunes-Nesi, A.; Williams, T.; Fernie, A.R. The mitochondrion: An integration point of cellular metabolism and signalling. Crit. Rev. Plant Sci. 2007, 26, 17–43.
- Jaipargas, E.A.; Barton, K.A.; Mathur, N.; Mathur, J. Mitochondrial pleomorphy in plant cells is driven by contiguous ER dynamics. Front. Plant Sci. 2015, 6, 783.
- Fuchs, P.; Rugen, N.; Carrie, C.; Elsässer, M.; Finkemeier, I.; Giese, J.; Hildebrandt, T.M.; Kühn, K.; Maurino, V.G.; Ruberti, C.; et al. Single organelle function and organization as estimated from Arabidopsis mitochondrial proteomics. Plant J. 2020, 101, 420–441.
- Sheahan, M.B.; Rose, R.J.; McCurdy, D.W. Organelle inheritance in plant cell division: The actin cytoskeleton is required for unbiased inheritance of chloroplasts, mitochondria and endoplasmic reticulum in dividing protoplasts. Plant J. 2004, 37, 379–390.
- Preuten, T.; Cincu, E.; Fuchs, J.; Zoschke, R.; Liere, K.; Börner, T. Fewer genes than organelles: Extremely low and variable gene copy numbers in mitochondria of somatic plant cells. Plant J. 2010, 64, 948–959.
- Arimura, S.I.; Yamamoto, J.; Aida, G.P.; Nakazono, M.; Tsutsumi, N. Frequent fusion and fission of plant mitochondria with unequal nucleoid distribution. Proc. Natl. Acad. Sci. USA 2004, 101, 7805–7808.
- Seguí-Simarro, J.M.; Coronado, M.J.; Staehelin, L.A. The mitochondrial cycle of Arabidopsis shoot apical meristem and leaf primordium meristematic cells is defined by a perinuclear tentaculate/cage-like mitochondrion. Plant Physiol. 2008, 148, 1380–1393.
- Unseld, M.; Marienfeld, J.R.; Brandt, P.; Brennicke, A. The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nat. Genet. 1997, 15, 57–61.
- Gualberto, J.M.; Mileshina, D.; Wallet, C.; Niazi, A.K.; Weber-Lotfi, F.; Dietrich, A. The plant mitochondrial genome: Dynamics and maintenance. Biochimie 2014, 100, 107–120.
- Gualberto, J.M.; Newton, K.J. Plant mitochondrial genomes: Dynamics and mechanisms of mutation. Annu. Rev. Plant Biol. 2017, 68, 225–252.
- Arimura, S.I.; Tsutsumi, N. A dynamin-like protein (ADL2b), rather than FtsZ, is involved in Arabidopsis mitochondrial division. Proc. Natl. Acad. Sci. USA 2002, 99, 5727–5731.
- Logan, D.C. Plant mitochondrial dynamics. Biochim. Biophys. Acta Mol. Cell Res. 2006, 1763, 430–441.
- Dyall, S.D.; Brown, M.T.; Johnson, P.J. Ancient invasions: From endosymbionts to organelles. Science 2004, 304, 253–257.
- Takanashi, H.; Arimura, S.I.; Sakamoto, W.; Tsutsumi, N. Different amounts of DNA in each mitochondrion in rice root. Genes Genet. Syst. 2006, 81, 215–218.
- Sheahan, M.B.; McCurdy, D.W.; Rose, R.J. Mitochondria as a connected population: Ensuring continuity of the mitochondrial genome during plant cell dedifferentiation through massive mitochondrial fusion. Plant J. 2005, 44, 744–755.
- Logan, D.C. The mitochondrial compartment. J. Exp. Bot. 2006, 57, 1225–1243.
- Arimura, S.I. Fission and fusion of plant mitochondria, and genome maintenance. Plant Physiol. 2018, 176, 152–161.
- Rose, R.J.; McCurdy, D.W. New beginnings: Mitochondrial renewal by massive mitochondrial fusion. Trends Plant Sci. 2017, 22, 641–643.
- Belliard, G.; Vedel, F.; Pelletier, G. Mitochondrial recombination in cytoplasmic hybrids of Nicotiana tabacum by protoplast fusion. Nature 1979, 281, 401–403.
- Rose, R.J.; Thomas, M.R.; Fitter, J.T. The transfer of cytoplasmic and nuclear genomes by somatic hybridisation. Funct. Plant Biol. 1990, 17, 303–321.
- White, R.R.; Lin, C.; Leaves, I.; Castro, I.G.; Metz, J.; Bateman, B.C.; Botchway, S.W.; Ward, A.D.; Ashwin, P.; Sparkes, I. Miro2 tethers the ER to mitochondria to promote mitochondrial fusion in tobacco leaf epidermal cells. Commun. Biol. 2020, 3, 161.
- Wakamatsu, K.; Fujimoto, M.; Nakazono, M.; Arimura, S.I.; Tsutsumi, N. Fusion of mitochondria in tobacco suspension cultured cells is dependent on the cellular ATP level but not on actin polymerization. Plant Cell Rep. 2010, 29, 1139–1145.
- El Zawily, A.M.; Schwarzländer, M.; Finkemeier, I.; Johnston, I.G.; Benamar, A.; Cao, Y.; Gissot, C.; Meyer, A.J.; Wilson, K.; Datla, R.; et al. FRIENDLY regulates mitochondrial distribution, fusion, and quality control in Arabidopsis. Plant Physiol. 2014, 166, 808–828.
- Lonsdale, D.M.; Brears, T.; Hodge, T.P.; Melville, S.E.; Rottmann, W.H. The plant mitochondrial genome: Homologous recombination as a mechanism for generating heterogeneity. Phil. Trans. Royal Soc. London B Biol. Sci. 1988, 319, 149–163.
- Bendich, A.J.; Gauriloff, L.P. Morphometric analysis of cucurbit mitochondria: The relationship between chondriome volume and DNA content. Protoplasma 1984, 119, 1–7.
- Kuroiwa, T.; Fujie, M.; Kuroiwa, H. Studies on the behavior of mitochondrial DNA: Synthesis of mitochondrial DNA occurs actively in a specific region just above the quiescent center in the root meristem of Pelargonium zonale. J. Cell Sci. 1992, 101, 483–493.
- Oldenburg, D.J.; Bendich, A.J. DNA maintenance in plastids and mitochondria of plants. Front. Plant Sci. 2015, 6, 883.
- Satoh, M.; Nemoto, Y.; Kawano, S.; Nagata, T.; Hirokawa, H.; Kuroiwa, T. Organization of heterogeneous mitochondrial DNA molecules in mitochondrial nuclei of cultured tobacco cells. Protoplasma 1993, 175, 112–120.
- Johnston, I.G. Tension and resolution: Dynamic, evolving populations of organelle genomes within plant cells. Mol. Plant 2019, 12, 7647–7683.
- Shen, J.; Zhang, Y.; Havey, M.J.; Shou, W. Copy numbers of mitochondrial genes change during melon leaf development and are lower than the numbers of mitochondria. Hortic. Res. 2019, 6, 95.
- Wolfe, K.H.; Li, W.H.; Sharp, P.M. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc. Natl. Acad. Sci. USA 1987, 84, 9054–9058.
- Palmer, J.D.; Herbon, L.A. Plant mitochondrial DNA evolved rapidly in structure, but slowly in sequence. J. Mol. Evol. 1988, 28, 87–97.
- Hu, J.; Huang, W.; Huang, Q.; Qin, X.; Yu, C.; Wang, L.; Li, S.; Zhu, R.; Zhu, Y. Mitochondria and cytoplasmic male sterility in plants. Mitochondrion 2014, 19, 282–288.
- Horn, R.; Gupta, K.J.; Colombo, N. Mitochondrion role in molecular basis of cytoplasmic male sterility. Mitochondrion 2014, 19, 198–205.
- Fitter, J.T.; Thomas, M.R.; Niu, C.; Rose, R.J. Investigation of Nicotiana tabacum (+) N. suaveolens cybrids with carpelloid stamens. J. Plant Physiol. 2005, 162, 225–235.
- Miyamura, S.; Kuroiwa, T.; Nagata, T. Disappearance of plastid and mitochondrial nucleoids during the formation of generative cells of higher plants revealed by fluorescence microscopy. Protoplasma 1987, 141, 149–159.
- Sato, M.; Sato, K. Maternal inheritance of mitochondrial DNA by diverse mechanisms to eliminate paternal mitochondrial DNA. Biochim. Biophys. Acta Mol. Cell Res. 2013, 1833, 1979–1984.
- Rice, D.W.; Alverson, A.J.; Richardson, A.O.; Young, G.J.; Sanchez-Puerta, M.V.; Munzinger, J.; Barry, K.; Boore, J.L.; Zhang, Y.; DePamphilis, C.W.; et al. Horizontal transfer of entire genomes via mitochondrial fusion in the angiosperm Amborella. Science 2013, 342, 1468–1473.




