Salvia miltiorrhiza is a renowned model medicinal plant species for which 15 SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) family genes have been identified; however, the specific functions of SmSPLs have not been well characterized as of yet. For this study, the expression patterns of SmSPL6 were determined through its responses to treatments of exogenous hormones, including indole acetic acid (IAA), gibberellic acid (GA3), methyl jasmonic acid (MeJA), and abscisic acid (ABA). To characterize its functionality, we obtained SmSPL6-ovexpressed transgenic S. miltiorrhiza plants and found that overexpressed SmSPL6 promoted the accumulation of phenolic acids and repressed the biosynthesis of anthocyanin. Meanwhile, the root lengths of the SmSPL6-overexpressed lines were significantly longer than the control; however, both the fresh weights and lateral root numbers decreased. Further investigations indicated that SmSPL6 regulated the biosynthesis of phenolic acid by directly binding to the promoter regions of the enzyme genes Sm4CL9 and SmCYP98A14 and activated their expression. We concluded that SmSPL6 regulates not only the biosynthesis of phenolic acids, but also the development of roots in S. miltiorrhiza.
1. Introduction
Salvia miltiorrhiza Bunge, which belongs to the Labiatae family, is a significant medicinal plant
[1]. Its dried roots (referred to as Danshen in Chinese), in combination with other herbs, have been used extensively for many years to treat various conditions
[2], including cardiovascular diseases
[3][4], menstrual disorders
[5], inflammation prevention
[6], hepatocirrhosis
[7], as an anti-osteoporotic
[2], and so on. Many capsules, dripping pills, injection solutions, and tablets used in clinical applications are comprised of Danshen.
S. miltiorrhiza contains diverse chemical components, encompassing approximately 50 diterpenoid quinones, more than 30 hydrophilic phenolic acids, and several essential oil constituents. According to pharmacological investigations, lipid-soluble tanshinones and water-soluble phenolic acids are the main active ingredients of
S. miltiorrhiza [1][8].
The biological activities of lipid-soluble tanshinones, such as tanshinone and tanshinol A, include cardio-cerebrovascular protection and serve as anticancer and anti-inflammatory agents
[9]. Hydrophilic phenolic acids, such as salvianolic acid B (SalB) and rosmarinic acid (RA), have potent anti-oxidative, anti-coagulation, and anti-inflammatory properties
[10]. As the main active ingredient of phenolic acids, SalB is designated as a primary component of Danshen in the official Chinese Pharmacopoeia
[11]. In
S. miltiorrhiza, the biosynthesis of RA and SalB originates from phenylpropanoid and tyrosine-derived pathways
[12]. The phenylpropanoid pathway includes three key enzymes, namely phenylalanine ammonia lyase (PAL), cinnamate 4-hydroxylase (C4H), and 4-coumarate-CoA ligase (4CL). For the tyrosine-derived pathway, 4-hydroxyphenylpyruvate reductase (HPPR) and tyrosine aminotransferase (TAT) are the key enzymes. Furthermore, rosmarinic acid synthase (RAS) and cytochrome P450 monooxygenase C3′H (CYP98A14) catalyze the biosynthesis of RA and SalB
[13][14][15]. Many reports have indicated that several transcription factors participate in the regulation of phenolic acid accumulation in
S. miltiorrhiza. For instance,
SmbHLH3 is a negative regulatory factor in the biosynthesis of phenolic acids
[16]. In contrast, the expression of
SmMYB111 can positively regulate the accumulation of phenolic acids
[17].
The
SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (
SPL) transcription factor family is characterized by a highly conserved SBP-box domain, which plays important roles in plant growth and development
[18]. The SBP-box domain consists of 76 amino acids that contain two zinc finger sites, which specifically bind to the GTAC core motif
[19].
SPL genes have been widely investigated in many plants. There are 16
SPL genes in
Arabidopsis thaliana [19], 18
SPL genes in
Betula luminifera [20], 19
SPLs in rice
[21], and 15
SPLs have been identified for
S. miltiorrhiza [22].
In
Arabidopsis, the functions of
SPL genes have been thoroughly investigated. Most
SPLs are miR156 targets, which is conserved and age-regulated in microRNA. The miR156-SPL regulatory module controls multiple developmental processes, including the juvenile-to-adult phase transition
[23], flower formation
[24], and root development
[25][26][27]. It has been reported that
AtSPL9 and
AtSPL15 mediate lateral bud growth and branching
[28].
AtSPL9 might interact with DELLA proteins (GA signaling pathway repressors) to promote the initiation of
Arabidopsis axillary buds
[29][30].
AtSPL9 also interacts with JAZ proteins and contributes to insect resistance in young plants
[31], whereas
AtSPL9 is involved in controlling the innate immunity of
A. thaliana [32]. Furthermore,
AtSPL9 prevents the expression of anthocyanin biosynthetic genes from down-regulating anthocyanin accumulation by directly interfering with the formation of a MYB-bHLH-WD40 transcriptional complex
[33].
Although 15 members of the
SPL family have been identified in
S. miltiorrhiza [22], none of these have been functionally experimentally characterized to date. According to phylogenetic tree analysis,
S. miltiorrhiza SPL6 (
SmSPL6) and
AtSPL9 tend to cluster in the same subgroup
[22]. We speculated as to whether
SmSPL6 might be involved in the accumulation of active ingredients in
S. miltiorrhiza. To investigate the functionality of
SmSPL6, we characterized its expression patterns and gain-of-function phenotype. Here, we found that
SmSPL6 responded to treatments with the exogenous hormones indole acetic acid (IAA), gibberellic acid (GA
3), methyl jasmonic acid (MeJA), and abscisic acid (ABA). The overexpression of
SmSPL6 promoted the accumulation of RA and SalB by directly binding to the promoter regions of
SmCYP98A14 and
Sm4CL9 and activating their expression. Meanwhile,
SmSPL6 repressed the biosynthesis of anthocyanins and altered the phenotype of root systems. All of the results indicated that
SmSPL6 is a strong regulator of both secondary metabolites and root development.
2. Discussion
2.1. Function of SmSPL6 in Phenolic Acid Biosynthesis
Phenolic acids are an intense area of research in the secondary metabolism of
S. miltiorrhiza. Previous reports have shown that several elicitors influence the production of phenolic acids
[34]. These elicitors can be divided into two groups (biotic and abiotic), with the former containing both pathogenic and plant cell components
[35][36], and the latter including Ag
+ [37], MeJA
[6], SA
[38], etc. Elicitors can affect phenolic acid compounds via transcription factors, which activate or repress the expression of enzyme genes that are engaged in the phenolic acid biosynthetic pathway. It was reported that some MeJA-responsive transcription factors, such as
SmMYB97
[39],
SmMYB1
[40],
SmMYB111
[17],
SmbHLH148
[41], and
SmERF1L1
[42], regulated the biosynthesis of phenolic acids via binding directly to and activating the promoters of enzyme-encoding genes involved in the biosynthetic pathway. For instance,
SmMYB1 positively regulates the biosynthesis of SalB by directly activating the key enzyme gene
SmCYP98A14 [40].
We found that
SmSPL6 responded to the treatment of exogenous MeJA (
Figure 1B). Besides
AtSPL9, the homeotic gene of
SmSPL6 is involved in the regulation of secondary metabolites
[35]. We speculated that
SmSPL6 could be a candidate transcription factor for regulating the accumulation of phenolic acids in
S.
miltiorrhiza. To investigate whether
SmSPL6 mediates the biosynthesis of phenolic acid, we generated
SmSPL6-overexpressed transgenic
S.
miltiorrhiza plants. The concentrations of RA and SalB were significantly increased in the transgenic lines. Y1H and dual-luciferase assays indicated that
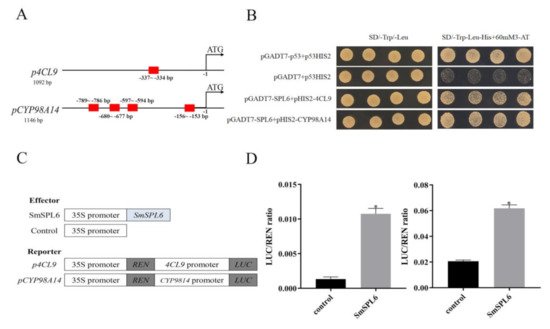
SmSPL6 could bind to the promoters of the key enzyme genes
SmCYP98A14 and
Sm4CL9 and activate their expression (
Figure 2). In this research, our findings demonstrated that
SmSPL6 was responsible for the generation of phenolic acid by directly activating the transcription of
SmCYP98A14 and
Sm4CL9 in
S.
miltiorrhiza.
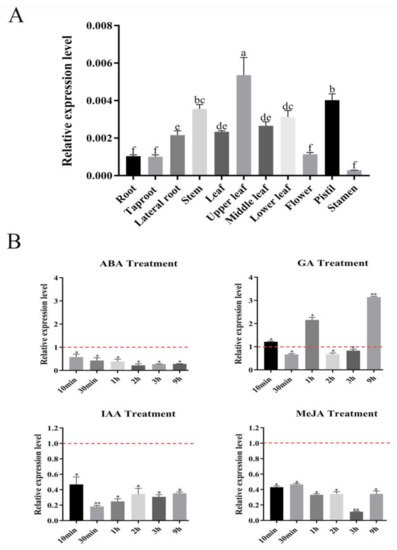
Figure 1. Expression profiles of SmSPL6 in Salvia miltiorrhiza. (A) The expression of SmSPL6 in different tissues. (B) Expression changes in response to treatment with 5 mM MeJA, 0.1 mM ABA, 0.1 mM IAA, and 0.1 mM GA3. All data are means of three biological replicates, with error bars indicating SD, red dotted line indicates the control which was set to 1. One-way ANOVA (followed by Tukey’s comparisons) tested for significant differences between means (indicated by different letters at p < 0.05). * and ** represent a significant difference at p < 0.05 and p < 0.01 compared with the control, respectively.
Figure 2. SmSPL6 binds to the promoter regions of Sm4CL9 and SmCYP98A14 and activates their expression. (A) GTAC motifs in the promoter regions of Sm4CL9 and SmCYP98A14. Red rectangles represent the GTAC motif. (B) Yeast one-hybrid detected interactions between the SmSPL6 and the promoters of Sm4CL9 and SmCYP98A14. The p53HIS2/pGADT7-p53 and p53HIS2/pGADT7 served as positive and negative controls, respectively. (C) Schematic diagram of constructs used in assays of transient transcriptional activity. (D) SmSPL6 activates the expression of Sm4CL9 and SmCYP98A14. Effector SmSPL6 was co-transformed with p4CL9-LUC/pCYP98A14-LUC reporters. All data are the means of three biological replicates, with error bars indicating SD; * represents a significant difference at p < 0.05 compared with the control.
2.2. Function of SmSPL6 in Anthocyanin Biosynthesis
Flavonoid-type anthocyanin is a critical type of secondary metabolite that can control plant fertility and protect plants from environmental stresses
[43][44][45]. It is also beneficial for human health due to its antioxidant, anti-cancer activities and anti-inflammatory function
[46][47]. Previous studies indicated that
AtSPL9 negatively regulates anthocyanin biosynthesis by disrupting the stability of the MYB-bHLH-WD40 transcription complex
[33]. Meanwhile,
AtSPL9 directly regulates the expression of
DFR (the key enzyme gene for anthocyanin biosynthetic pathway) to influence the metabolism of anthocyanin
[33]. We detected the content of anthocyanin in the
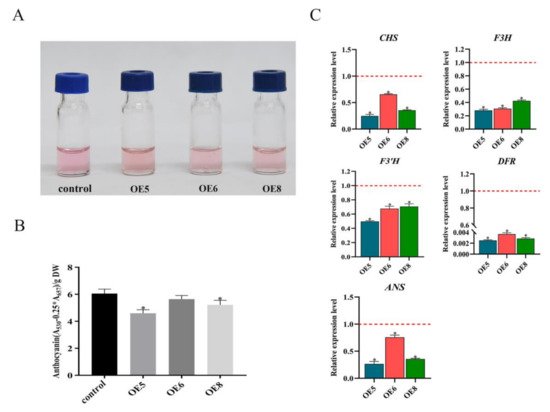
SmSPL6-OE transgenic lines and the control. Our results showed that overexpressed
SmSPL6 reduced the accumulation of anthocyanin in
S.
miltiorrhiza (
Figure 3), which was consistent with the function of
AtSPL9 in regulating the production of anthocyanin. The expression levels of enzyme-encoding genes for the anthocyanin biosynthesis pathway were all down-regulated in the
SmSPL6-OE lines (
Figure 3C). We analyzed the promoter regions of those genes and found that the GTAC motif existed in the promoter regions of
CHS,
F3H,
F3′H,
DFR, and
ANS (data not shown). Whether
SmSPL6 regulates the expression of these genes by directly binding to the GTAC motif will be investigated in our future studies.
Figure 3. SmSPL6 negatively regulates the biosynthesis of anthocyanin. (A) Anthocyanin extracted from the whole plant of the transgenic lines and the control. (B) Concentrations of anthocyanin in the transgenic lines and the control. (C) Expression changes of the enzyme genes for the biosynthetic pathway of anthocyanin. CHALCONE SYNTHASE (CHS), FLAVANONE 3-HYDROXYLASE (F3H), FLAVONOID 3′-HYDROXYLASE (F3’H), DIHYDROFLAVONOL REDUCTASE (DFR), ANTHOCYANIDIN SYNTHASE (ANS). All data are the means of three biological replicates, with error bars indicating SD. Red dotted lines indicate the control value, which was set to 1. * represents a significant difference at p < 0.05 compared with the control.
Water-soluble phenolic acids and anthocyanin share a common phenylpropanoid pathway. Previous literature indicated that the positive regulators of phenolic acids may have different roles to play in the production of anthocyanin. For instance, overexpressed
SmMYB1 significantly promoted the accumulation of anthocyanin
[40], while overexpressed
SmbHLH51 did not significantly alter anthocyanin generation
[48]. Our results indicated that
SmSPL6 was a positive regulator for phenolic acids, but a negative regulator for anthocyanin, revealing that the regulatory mechanisms of secondary metabolites in plants is quite complex.
2.3. Function of SmSPL6 in Root Development
Root systems are essential for plant growth and survival due to their critical roles in the acquisition of water and nutrients. As is well known, the dried roots of
S.
miltiorrhiza are used as a traditional Chinese medicine; thus, improving the biomass and quality of roots is an important goal for the breeding of
S. miltiorrhiza. Earlier reports have shown that
AtSPL9 and
AtSPL10 repressed lateral root growth in
Arabidopsis [27]; 10-day-old
pSPL9:rSPL9 seedings exhibited fewer lateral roots than the wild type, whereas
pSPL10:rSPL10 seedings exhibited the delayed generation of lateral roots in contrast to
pSPL9:rSPL9, which indicated that
AtSPL10 played a major role in lateral root growth
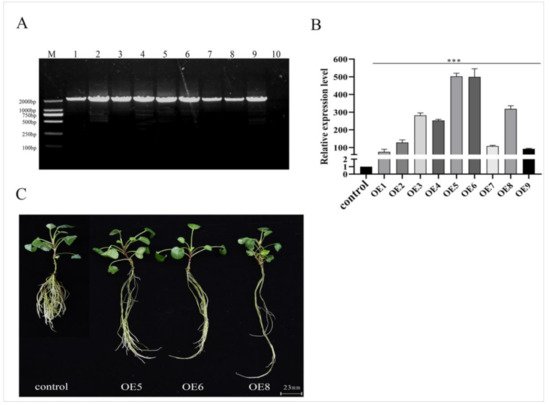
[49]. We observed obvious changes in the root phenotypes, including fewer lateral roots, longer root lengths, and wider root diameters in the
SmSPL6-OE lines (
Figure 4C and
Table 1). Although the root biomass decreased in the
SmSPL6-OE lines, the phenotype of fewer lateral roots and longer root lengths are preferred for this traditional Chinese medicinal material.
Figure 4. Identification and phenotype of the SmSPL6-overexpressed transgenic Salvia miltiorrhiza. (A) PCR-amplification product from the genome DNA of transgenic lines and the control. Forward primer: CaMV35S–F, reverse primer: SmSPL6–R, full length: 1833 bp. Lanes M, DL2000 DNA marker; 1–9, different transgenic lines; 10, control line obtained by plant tissue culture without Agrobacterium infection. (B) qRT-PCR analysis. Fold changes reflect the expressions of transgenic lines compared with the control expression, where the control values were set to 1. All data are the means of three replicates, with error bars indicating SD; *** represents a significant difference at p < 0.01 in contrast to the control. (C) Phenotype of the SmSPL6-overexpressed OE5, OE6, and OE8 transgenic lines and the control.
Table 1. Root parameters of the SmSPL6 overexpressed (OEs) transgenic lines and the control.
| Lines |
Roots Length/cm |
Fresh Weight/g |
Lateral Roots Number |
| control |
8.54 ± 0.87 c |
0.760 ± 0.082 a |
>20 a |
| OE5 |
16.02 ± 0.52 b |
0.625 ± 0.064 a |
15 ± 2 b |
| OE6 |
17.15 ± 0.49 b |
0.600 ± 0.028 a |
15 ± 2 b |
| OE8 |
20.10 ± 0.71 a |
0.385 ± 0.035 b |
12 ± 2 b |
Note: One-way ANOVA (followed by Tukey’s comparisons) tested for significant differences between means (indicated by different letters at p < 0.05).
The plant hormone auxin plays vital roles in the growth and development of roots
[50][51]. Whether
SmSPL6 inhibits lateral root development by regulating the levels of endogenous auxin should be further investigated for
S. miltiorrhiza. In
Arabidopsis, the expression of
AtSPL9 and
AtSPL10 was induced through the treatment of exogenous IAA
[49]. Our data indicated that
SmSPL6 was responsive to auxin; however, its expression was inhibited by the exogenous IAA treatment (
Figure 1B). The opposite expression responses of
SmSPL6 and
AtSPL9 to IAA may have been due to the application of different concentrations of exogenous IAA. In the present study, 100 μM IAA was used to spray the
S. miltiorrhiza seedlings, while the
Arabidopsis seedlings were treated with 10 μM IAA. Whether
SmSPL6 is induced by low concentrations of IAA will be further investigated.
Collectively, these results elucidated the role of SmSPL6 in the regulation of secondary metabolites and lateral root development in S. miltiorrhiza. The functional consistency of SmSPL6 and AtSPL9 for inhibiting lateral root development and the biosynthesis of anthocyanin revealed the conservatism of the SPL family in plants, while the function of SmSPL6 in promoting the generation of SalB demonstrated the species specificity of SPL members. In the following research, we will try to generate SPL6 mutant lines in S. miltiorrhiza using the CRISPR/Cas9 system to better elucidate the function of SmSPL6 transcription factor.