Video Upload Options
Antibiotics are defined as natural or synthetic chemicals inhibiting both the growth and survival of microorganisms. These compounds have been used as essential therapeutics for nearly a century. With the human population growth increasingly, antibiotics become preventive agents in the agricultural and animal industry. The multi-faceted global use and misuse of antibiotics led to the evolution of bacteria into antibiotic resistant species. Advantageous mutations cause antibiotic tolerance, which is transmitted to offspring (vertical evolution) or to another bacteria via conjugation, transduction, or transformation mode (horizontal evolution), that are then are then passed down to progeny (vertical evolution). Antibiotic resistance progressively increased mortality from multidrug-resistant bacterial infections, threatening public health. Probiotics, essential oils, vaccines, and antibodies to antibiotic resistant bacteria emerge as primary or adjunct preventive measures or therapies against multidrug-resistant bacterial infections. Bacteriophages and predatory bacteria, as well as utilization of bacterial communication quorum sensing considered as new ecosystem-influenced methods to tackle multidrug-resistant bacteria. Mechanisms of dissemination and the development of antibiotic resistance genes vary in the context of nutrition and related clinical, agricultural, veterinary, and environmental settings, this need to be accounted in selections of preventive and therapeutic countermeasures.
Antibiotic resistance is the process in which fungi or bacteria gain the ability to tolerate toxic influence, adapt and proliferate in the presence of antibiotics. Antibiotic resistance poses a threat to human and animal health and challenges the agricultural food supply and the integrity of the environment worldwide. Relentless continued use and misuse of antibiotics created a continuum for perpetuate horizontal and vertical evolution of antibiotic resistant bacteria proliferating in unfavorable environments of newer and more antibiotics. This continuum makes classification required to document all antibiotic resistance threats not be feasible. Although antibiotics are utilitarian part of modern medicine for nearly century, their efficacy and safety do not meet the demands of growing population facing global threat of antibiotic-resistant infectious diseases. Similar challenge encounters intensifying animal production. The complex global antibiotic resistance phenomena required innovative multilayered interdisciplinary solutions. Four major strategic directions are developing: 1) utilizing immune response with immune therapies, antibodies and plant-based metabolites, stimulating immunity along with antibiotic effects; 2) Physical devices, including hemofiltration devices; 3) antimicrobial adjuvants (AA) and nitric oxide-based nanotherapies disrupting bacterial pathways; and 4) ecosystem-influenced methods, utilizing predatory bacteria, bacteriophage, transplantation of fecal microbiota, and quorum sensing inhibitors, intercepting bacterial communication. Combating global spread of antibiotic resistance also requires comprehensive and incessant efforts of policymakers in agriculture working with experts in diverse fields of microbiology, biochemistry, clinical research, genetic, and computational engineering.
1. Introduction
Since penicillin discovery by Alexander Fleming [1] (Figure 1) and its extraction from a fungus Penicillium chrysogenum, by Howard Florey and Ernst Chain in 1941, penicillin, the most effective, methicillin and other antibiotics are produced in copious amounts. The advances in social and medical fields, including cancer therapy and organ transplantation, would not have been possible without effective antibiotic treatment to control bacterial infections. The term antibiotic is defined as a natural or synthetic chemical inhibiting both the growth and survival of microorganisms. However, the emergence of methicillin resistant S. aureus (MRSA) coincide with reported increase in sepsis cases from 621,000 to 1,141,000 between the years of 2000 and 2008 [2]. The death toll from sepsis rose from 154,000 to 207,000 cases. MRSA marks the beginning of the development of antibiotic–resistant microbes (also called ESKAPE pathogens, standing for Enterococcus faecium, S. aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacteriaceae) [3]. It is reported that in the United States, India, Thailand, and European Union, antibiotic resistance causes more than 23,000, 58,000, 38,000 and 25,000 deaths per year, respectively [4–7]. The predicted deaths from drug-resistant microbial pathogens could rise from approximately 700,000 per year to 10 million deaths per year by 2050 and threaten global health [8].
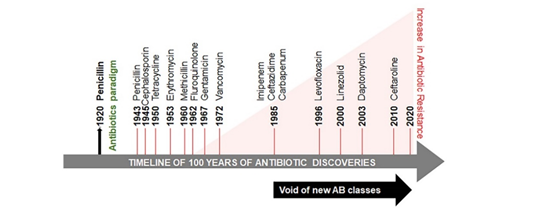
Figure 1. Timeline of antibiotic discovery and its onset of resistance. The antibiotic paradigm emerges out of the followed discovery of penicillin. Between the 1960s and the 1980s there was a surge in the discovery of antibiotics, but this development declined between the 1980s and the 1990s. The identification of new antibiotic classes by pharmaceutical companies has stagnated since 1987 and coincided with progressively increased antibiotic resistance and mortality related to antibiotic-resistant infections.
Microorganisms are able to develop antibiotic-resistant genes to enhance their survival, thus minimizing the treatment options for microbial infections and increasing mortality in human populations. Antibiotic resistance is classified into three categories based on the threat: urgent, serious, and concerning (Table 1). The global threat of resistance to imipenem antibiotics in A. baumannii infections has been reported in both Organization for Economic Co-operation and Development (OECD) and non-OECD countries across the globe [9]. Several reasons are responsible for the development of antibiotic resistance globally and in developing countries, such as India [10,11]. Poor public health conditions and health care systems, availability of antibiotics over the counter, lack of public knowledge of appropriate dosage of antibiotics and their haphazard use, as well as a high incidence of infectious diseases have been proposed as the major factors augmenting the problem. This continuum of antibiotic resistance concept was proposed to describe the progressive interconnecting influence of human, industrial, agricultural, and wild environments [12]. The crude mortality due to infectious diseases in India is 416.75 per 100,000 persons, which is twice the rate in the United States (roughly 200 per 100,000 persons) [13]. The problem is aggravated further by the void in the development of new classes of antibiotics since 1990 (Figure 1) [14–17].
Table 1. Classification of antibiotic resistance threats.
|
Urgent |
Serious |
Concerning |
|
1. A. baumannii, P. aeruginosa, carbapenem-resistant 2.Clostridium difficile (CDIFF) 3. N. gonorrhoeae-3rd generation cephalosporin-resistant, fluoroquinolone-resistant 4. Carbapenem- and 3rd generation cephalosporin resistant Enterobacteriaceae: K. pneumonia, E. coli, Enterobacter spp., Serratia spp., Proteus spp., and Providencia spp., Morganella spp. |
1.Streptococcus pneumonia, penicillin-non-susceptible 2. Haemophilus influenzae, ampicillin-resistant 3. Shigella spp., fluoroquinolone-resistant 4. Enterococcus spp., vancomycin resistant 5. Multidrug-resistant Acinetobacter 6. Drug resistant Campylobacter 7.Extended-spectrum β-lactamase producing Enterobacteriae (ESBLs) 8. Multidrug-resistant P. aeruginosa 9. Drug-resistant non-typhoidal Salmonella 10. Drug-resistant Salmonella serotype Typhi 11. Drug resistant M. tuberculosis 12. Methicillin-resistant S. aureus (MRSA) |
1. Group B Streptococcus (GBS), clindamycin resistant 2. Group A Streptococcus (GAS), erythromycin resistant 3. S. aureus, vancomycin resistant |
Evidence of various factors leading to the development of antibiotic resistance, followed by route of entry of drug-resistant pathogens into the food chain, and a plethora of alternative strategies to mitigate the menace of antibiotic resistance for a healthier future is reviewed .
2. Drug Resistance Continuum
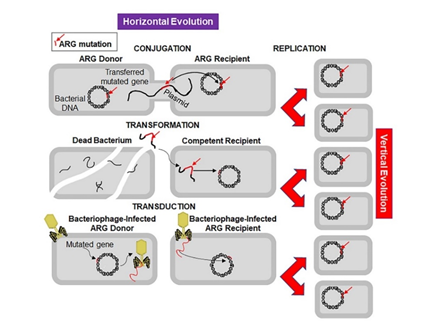
Antibiotic resistance appeared soon after clinical use of antibiotics, because microorganisms are evolving rapidly to endure and proliferate in unfavourable environments. Sulphonamide-resistant Streptococcus pyogenes appeared in the human clinical settings in early 1930s, while penicillin-resistant S. pyogenes was noted in the 1940s. The problem of multidrug-resistant enteric bacteria became noticeable in the 1950s [19]. Antibiotic resistance develops as a result of transmittion of advantageous mutations to progeny (vertical evolution) or to another bacteria via conjugation, transduction, or transformation mode (horizontal evolution) (Figure 2). Human pathogens use horizontal gene transfer is an important mechanism of antibiotic resistance gene (ARG) acquisition among microorganisms along with the vertical transfer [20]. A decade ago, an ARG, the New Delhi metallo-β-lactamase 1 (NDM-1) was identified in single isolates of K. pneumonia and Escherichia coli in a patient in New Delhi, India, who’s repatriatiation to Sweden [21] led to the spread of antibiotic resistance worldwide [22,23]. NDM-1 has no detectable sequence homology with other classes of these genes, thus indicating their archaic origin [24,25]. Bacteria carrying extended spectrum β-lactamases (ESBL) impart resistance to penicillin and cephalosporins, extensively drug-resistant (XDR) Mycobacterium tuberculosis, and multi-drug resistant A. baumanni, Enterobacteriaceae, Neisseria gonorrhoea, and P. aeruginosa [26,27].
Figure 2. Mechanisms of horizontal and vertical transmission in bacteria for the development of antibiotic resistance. The left panel shows the horizontal transmission of an antibiotic resistant gene (ARG, red line indicated by a red arrow) by the three main mechanisms: conjugation, transformation, and transduction. Conjugation involves transfer of the ARG from a donor bacterium to a recipient by direct contact and plays a crucial role in dissemination of antibiotic resistance. Transformation involves uptake of the free DNA with the ARG from the environment. Transduction is a virus-mediated gene transfer by bacteriophages. The right panel shows the vertical evolution carried out by replication of bacteria containing the ARG.