| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Federico Lazzaro | + 2770 word(s) | 2770 | 2020-03-11 02:59:37 | | | |
| 2 | Camila Xu | -24 word(s) | 2746 | 2020-11-01 14:38:44 | | |
Video Upload Options
RNA is frequently found into DNA. Single embedded ribonucleotides are mainly introduced by DNA polymerases. Longer stretches of RNA can also anneal to DNA, forming RNA:DNA hybrids, as occurs for R-loops. Even if R-loops are the most studied hybrid structures, the world of RNA:DNA hybrids is much wider. Polyribonucleotide chains are indeed synthesized to allow Okazaki fragments priming in the process of DNA replication, and double-strand breaks repair and may also result from the direct incorporation of several consecutive ribonucleotides by DNA polymerases. We discuss about all the possible sources of single and multiple ribonucleotides in DNA, focusing on situations where the aberrant processing of RNA:DNA hybrids may result in potentially harmful stretches of consecutive ribonucleotides embedded into the genome, whose existence is also supported by their presence into the DNA of organelles.
1. Single embedded ribonucleotides are introduced in the genome by DNA polymerases
Most leaving organisms store their genetic information in DNA rather than in RNA, due to the inherent chemical instability of the RNA. The DNA, indeed, lacks the reactive 2ʹ-OH group on the ribose sugar, which can attack the sugar-phosphate backbone, generating breaks with genotoxic outcomes [1]. Apart from choosing the proper complementary base, replicative DNA polymerases must then discriminate between ribose in rNTPs versus deoxyribose in dNTPs [2]. This is why they possess a special “steric-gate” residue in their nucleotide-binding pocket. Steric-gate residues, like Tyrosine or Phenylalanine in replicative polymerases, are characterized by a bulky side chain that sterically clashes with the 2’-OH of incoming rNTPs, preventing their incorporation in DNA [3]. Other active site residues contribute to keeping the side chain of the steric-gate residue and the incoming nucleotide in the proper orientation to achieve high sugar selectivity [3]. Moreover, it was recently shown that a polar filter interacts with the 3’-OH and the triphosphate moiety of the incoming nucleotide; this causes the 2’-OH of a rNTP to clash with the surface of the fingers domain, further limiting the possibility to bind rNTPs in a catalytically competent conformation [4].
Sugar selectivity, however, is not perfect, especially because DNA polymerases are constantly challenged by high rNTP concentrations. For example, even if in yeast cells the dNTP pools increase of about three-fold upon entry into the S phase respect to G1 [5], the physiological concentrations of the four rNTPs greatly exceed those of dNTPs: rNTPs range from 500 to 3,000 μM, while dNTPs are in between 12 and 30 μM [6]. For this reason, pol ε was estimated to introduce 1 rNMP every 1,250 dNMPs during leading strand synthesis, while pol δ and pol α insert of 1 rNMP every 5,000 dNMPs and 625 dNMPs respectively, during lagging strand synthesis [6]. This results in more than 13,000 rNMPs inserted into the yeast genome for each replication cycle [6]. Such high numbers also depend from the reduced ability of pol ε and especially pol δ to proofread rNMPs [7][8][9]. Ribonucleotides can be considered as the most common non-canonical nucleotides present in the eukaryotic genome [6][10]. The presence of ribonucleotides into genomic DNA was confirmed in vivo by alkali-sensitivity assays [11]. Single or di-ribonucleotides were detected in vivo also in mammalian genomic DNA and estimated to generate at least 1,000,000 alkali-sensitive sites per cell [12].
The activity of pol ε and pol δ is not restricted to DNA replication. They also work in repair processes requiring DNA synthesis, in particular nucleotide excision repair (NER) [13], so they may also introduce rNMPs in such circumstances. Reparative DNA synthesis steps are as well performed by many other specialized polymerases that can contribute to rNMPs incorporation [14][15]. It should be emphasized that these polymerases are often active outside of the S phase [16][17][18][19], when the concentration of dNTPs is even lower than in the S phase, which may contribute to a more significant incorporation of rNMPs into DNA. We can then speculate that “non-replicative” ribonucleotides may become particularly relevant in post-mitotic cells, such as neurons, where TLS was recently found to take place [20].
2. Main mechanisms of single ribonucleotides removal
To maintain the correct DNA:DNA composition cells have evolved ribonucleases H (RNases H) specialized in the removal of ribonucleotides from DNA. In eukaryotic cells, RNase H2 is composed by three subunits (Rnh201, Rnh202, Rnh203 in yeast; RNaseH2A, RNaseH2B, RNaseH2C in higher eukaryotes) all essential for the activity of the complex, and it cleaves both single and multiple rNMPs paired with DNA [21]. RNase H2 is the initiator of ribonucleotide excision repair (RER), the most common repair pathway for removal of genomic embedded rNMPs [10]. RER ensures genome integrity and proper development of mouse embryos [12], keeping embedded rNMPs under a threshold of ribonucleotide tolerance [22]. RNase H2 mutations are associated with a rare autoinflammatory disorder known as Aicardi-Goutières syndrome (AGS)[23], mainly characterized by early-age onset, and chronic overproduction of type I interferon in the absence of infections [24]. Patient-derived cells accumulate rNMPs in their genome and exhibit constitutive post-replication repair (PRR) and DNA damage checkpoint activation [25][26].
Eukaryotic cells also possess RNase H1, which is a single subunit protein that cleaves stretches of at least 4 consecutive rNMPs [21]. Its enzymatic activity is essential for mitochondrial DNA replication in mammals [27], while it does not seem to be required during RER [10].
3. Possible sources of multiple embedded ribonucleotides in the genome
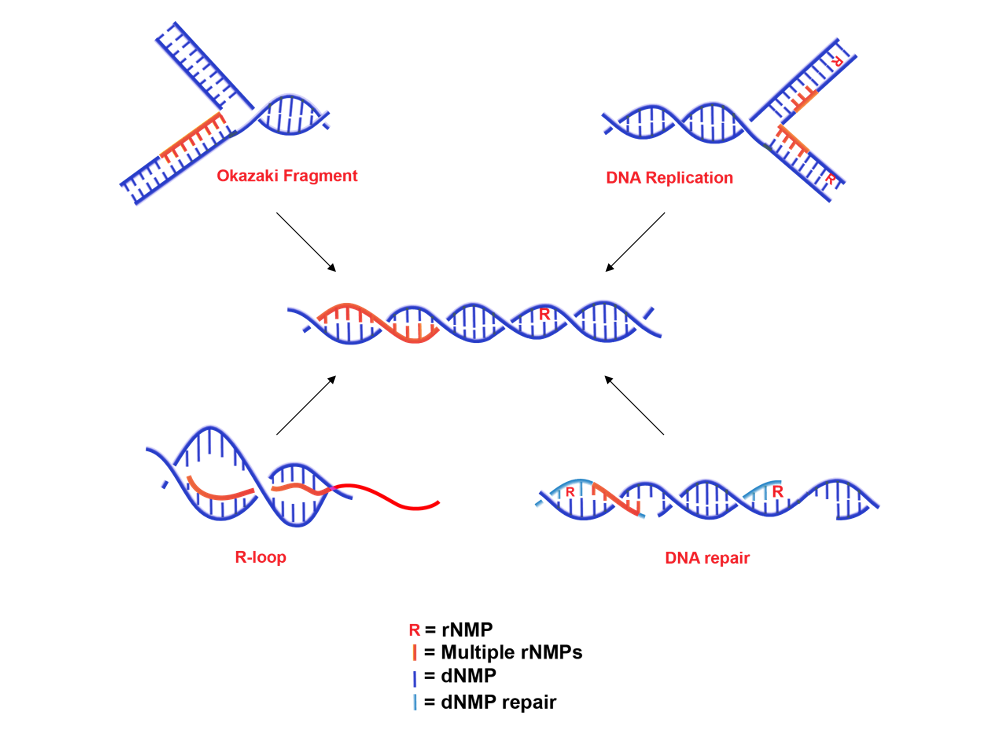
Although the presence of single ribonucleotides into the chromosomal DNA has been extensively investigated in different organisms, whether the incorporation of consecutive rNMPs is also possible is still unclear. The study of multiple embedded rNMPs is complicated by the fact that it requires the simultaneous removal of RNase H1 and H2, which can both recognize stretches of more than 4 consecutive rNMPs. S. cerevisiae is an excellent model organism to this purpose, because mutants lacking all RNase H activities are still viable [28]. Nevertheless, RNases H can potentially process any polyribonucleotide tract in DNA (stretches of rNMPs, R-loops, RNA primers found at Okazaki fragments, etc.), so it remains difficult to establish which one of these unprocessed substrates causes the observed effects. Anyway, if stretches of consecutive rNMPs do exist, how they are incorporated (Figure 1) and subsequently removed, needs to be clarified. We discuss below different possible sources of multiple embedded rNMPs.
Figure 1. Sources and forms of rNMPs embedded in genomic DNA.
Despite DNA polymerases being primarily responsible for the incorporation of single rNMPs, only mutant variants seem capable of introducing consecutive rNMPs. The pol ε variant pol2-M644G incorporates rNMPs in DNA at higher frequencies than the wild type pol ε [11]. The fact that this mutant becomes synthetic lethal with the simultaneous absence of RNase H1 and H2 suggests that it incorporates stretches of rNMPs, requiring the activity of both RNases H to be removed [28][29]. Also the S. cerevisiae polη-F35A steric-gate mutant seems to incorporate polyribonucleotide tracts in DNA at a high rate, leaving a specific 1bp deletion signature without RNase H2 [30][31]. Moreover, under particular stress conditions, also wild type replicative and/or reparative DNA polymerases may incorporate consecutive rNMPs. For example, Meroni et al. found that, upon replication stress induced by hydroxyurea, pol η is recruited at stalled replication forks, where it facilitates the formation of stretches of rNMPs that become highly toxic for cells, when not replaced by DNA [32].
Although the number of rNMPs incorporated during DNA replication is surprisingly large, the main source of genomic ribonucleotides remains by far the replication priming. Replicative DNA polymerases require a piece of RNA initiator (RNAi) of ~ 8-10nt in length to work. Considering the discontinuous nature of the lagging strand, this results in about 100,000 RNA:DNA hybrids in yeast and in more than 10 millions of hybrids in human cells, for each round of DNA replication [33][34]. RNA primers must then be removed, and Okazaki fragments (OFs) joined together to form a continuous lagging strand. Different pathways cooperate in Okazaki fragments maturation [35]; the dominant one depends on Fen1 (Rad27 in S. cerevisiae) [36][37][38]. In the absence of strand displacement, also RNase H2 seems to have a role in the direct hydrolysis of RNA:DNA primers [39]. S. cerevisiae strains lacking Rad27 and RNase H2 are sick, becoming lethal when combined with RNase H1 deletion. This suggests that, besides RNase H2, also RNase H1 has a role in Okazaki Fragments maturation [40]. The exact composition, crosstalk, and regulation of all these pathways are still largely unknown, but dysfunctions in any of these mechanisms could leave flaps or nicks into the genome, causing deletions, amplification of DNA sequences, and DSBs [41]. Moreover, even if never verified, dysfunctions could also result in the stable inclusion of RNA stretches into DNA, as suggested by different groups [32][42]. Intriguingly, Holmes et al. [43] found that this happens in the mitochondrial DNA of mice, where, in the absence of RNase H1, RNA primers are retained in both template strands, causing dramatic effects on mtDNA replication. The incorporation of an RNA primer into the DNA is also the mechanism proposed for mating-type switching in S. pombe. During the S phase, two consecutive rNMPs are left by incomplete processing of RNA primer into the lagging strand at the MAT1 locus; these rNMPs are maintained until the following replication cycle, inducing polymerase stalling, and recombination events, which lead to mating-type switching [44][45].
Another important source of ribonucleotides is represented by R-loops, peculiar three-stranded structures formed when a transcribed RNA hybridizes back to the template, leaving the non-template DNA single-stranded [46]. These hybrid regions are longer than the canonical 8bp hybrids formed by active RNA polymerases (RNAPs) [47] and R-loop-prone regions cover about 8% of the yeast genome [48]. Growing evidence suggests that these structures play important roles in regulating gene expression [49] and chromatin structures [50]. On the other hand, they can compromise genome integrity since R-loops expose patches of ssDNA, which are more susceptible to mutagenesis, recombination and DNA damage, compared to dsDNA [51]. Moreover, conflicts between the DNA replication machinery and R-loops trigger fork collapse and DSBs [52][53]. Once formed, different factors can act to remove R-loops, like RNase H enzymes (H1 and H2), which cleave the RNA moiety of RNA:DNA hybrids [21] and numerous helicases, that unwind the hybrids, such as Senataxin (Sen1 in S. cerevisiae) [54], the human DHX9 [55] and Pif1-family helicases [56]. However, different situations have been described where the RNA filaments of R-loops become embedded into DNA. In prokaryotic cells R-loops are frequently associated with origin-independent replication [57][58]. In vitro studies have shown that prokaryotic DNA polymerases can use mRNA as a primer when the replication fork collides with the RNA polymerase [59], and this is also the case for eukaryotic cells. Stuckey et al. [60] found that in S. cerevisiae RNA polymerase I transcription constraints lead to persistent R-loops in ribosomal DNA, where the RNA present in the R-loop can be used as a primer by DNA polymerases, triggering an origin-independent replication process. Being highly inaccurate, this unscheduled replication can cause genome instability.
Finally, the local incorporation of ribonucleotides, and the presence of different types of RNA molecules have been shown to have important effects even on DNA DSBs, influencing their repair by nonhomologous end-joining (NHEJ) or homologous recombination (HR) pathways [61][62][63]. For example, Pryor et al. recently reported that one or more rNMPs are transiently incorporated at broken DNA ends by pol μ or TdT, enhancing DSB repair by NHEJ mechanisms [64]. Growing evidence shows that also the hybridization of complementary RNA molecules at DSB ends regulates their repair [61][62][63][65]. RNA:DNA hybrids seem to contribute to the recruitment of repair factors [66][67][68][69][70][71][72][73][74][75], and to the control of DNA end resection [67][72][74][75], the fundamental process creating 3ʹ end ssDNA filaments needed for recombination [76]. However, how RNA:DNA hybrids impact on DSBs processing and repair is still an open debate [77]. Indeed, while some data indicate that they promote resection [71][72][75], others suggest an anti-resection role [67][74] or no effect at all [69]. More work is thus required to clarify the regulation of this dynamic phenomenon, also because, as mentioned for R-loops, and RNA primers at Okazaki fragments, it is tempting to speculate that improperly removed RNA tracts might remain embedded at DSB ends, posing a threat to genome stability.
4. Mechanisms of Multiple Embedded Ribonucleotides Removal
As previously mentioned, single rNMPs are the substrate of RER [10], but whether this pathway works on multiple rNMPs has never been proved. It is unlikely that the pathways acting on R-loops and OFs could process multiple rNMPs, once embedded into DNA, and thus inaccessible to players like helicases. Since RNase H1 and H2 both process consecutive embedded rNMPs, they represent the main candidates for their removal. Anyway, how the two enzymes work in vivo on these structures needs further clarification. Some progress has been made thanks to the development of a separation-of-function mutant of the RNase H2 enzyme, called rnh201-RED (ribonucleotide excision defective), which loses the ability to remove single rNMPs, but retains a discrete activity on consecutive rNMPs [78]. This mutant has been extremely useful to enlighten the role of the two functions of RNase H2 [79]. Being still able to remove multiple rNMPs, the rnh201-RED mutant alone cannot prove their existence; the development of additional separation-of-function mutants may thus be useful.
5. Multiple ribonucleotides into the DNA of organelles
Multiple ribonucleotides have also been detected into the DNA of two types of eukaryotic organelles: mitochondria and chloroplasts.
The human mitochondrial DNA (mtDNA) is a circular multicopy molecule of 16,5kb, composed of two filaments, named heavy (H) strand and light (L) strand, and whose replication mechanism is not completely resolved. Different models for mitochondrial DNA duplication have been proposed [80][81]. Replication primers represent the first source of consecutive rNMPs also in mtDNA. They seem to be synthesized by the mitochondrial RNA polymerase POLRMT and not by a replicative primase, as for the nuclear DNA [82]. Such transcripts are stabilized by G-quadruplex structures formed in the non-template DNA strand, resulting in mitochondrial R-loops that act as replication primers [83]. Polyribonucleotide chains could also result from long RNA transcripts, which temporally coat the displaced H-strand, generating RNA:DNA hybrids that function as lagging strands during mtDNA replication. These long RNAs may result from a primase activity or by the hybridization of the displaced DNA with preformed RNA transcripts [84]. RNase H1 is the factor responsible for the removal of multiple rNMPs from mtDNA. The mammalian RNase H1 is recruited into the organelles thanks to an essential mitochondrial localization domain, and failures in its activity cause mitochondrial dysfunctions. In mouse, when RNase H1 is absent, replication primers are not properly removed and stretches of RNA remain fixed in both template strands of mtDNA [43]. Since they cannot be bypassed by the mtDNA polymerase γ, they lead to persistent DNA gaps that are catastrophic for the subsequent round of replication [43]. As a consequence, mice lacking RNase H1 die during embryogenesis [27]. In human, mutations in RNase H1 have been associated with mitochondrial encephalomyopathy with adult onset [85]. These examples highlight the importance of removing multiple rNMPs from mtDNA.
Ribonucleotides have also been observed into the DNA of chloroplasts, the other organelles capable of autonomous replication in plant cells. The chloroplast DNA (cpDNA) consists of linear or circular multicopy molecules of 120-170kb, which can replicate in different manners [86]. Even if there is still much to learn about rNMPs in the DNA of chloroplasts, it is evident that stretches of multiple rNMPs can compromise cpDNA stability. Apart from RNA tracts used for DNA replication priming, R-loops can be frequently found in these organelles. It has been found that the AtRNaseH1-like protein (RNH1C), together with DNA gyrases, plays a key role in the processing of these hybrids, maintaining chloroplast DNA integrity [87][88].
Even if rNMPs in mtDNA and cpDNA need to be further explored, their existence in these endosymbiotic organelles is extremely intriguing: it suggests that the presence of the incorrect sugar in DNA comes from ancestral forms of life and has been maintained throughout the evolution, to perform physiological functions also in evolved organisms.
6. Concluding Remarks
Although stretches of multiple embedded rNMPs have only been observed in mtDNA, their presence into the nuclear DNA has also been genetically predicted. The persistence of multiple rNMPs in the mitochondrial DNA has been shown to have detrimental effects, and so is suspected for genome-embedded polyribonucleotide chains, with consequences even more severe than those deriving from unprocessed single rNMPs. Different techniques are currently available to study single rNMPs and RNA:DNA hybrids, but further efforts should be made for the development of groundbreaking methods allowing to isolate only the desired category of RNA:DNA hybrids, and to distinguish sites of single rNMPs insertion from sites with multiple rNMPs. Demonstrating the existence of consecutive embedded rNMPs and discovering details about their sources and removal might help to clarify the contribution of the two RNases H in the recognition and processing of all hybrid structures, and, importantly, to shed light on the mechanisms linking RNA:DNA hybrid structures, replication stress, genome instability and severe human pathologies.
References
- Yingfu Li; Ronald R. Breaker; Kinetics of RNA Degradation by Specific Base Catalysis of Transesterification Involving the 2‘-Hydroxyl Group. Journal of the American Chemical Society 1999, 121, 5364-5372, 10.1021/ja990592p.
- C. M. Joyce; Choosing the right sugar: How polymerases select a nucleotide substrate. Proceedings of the National Academy of Sciences 1997, 94, 1619-1622, 10.1073/pnas.94.5.1619.
- Jessica A. Brown; Zucai Suo; Unlocking the Sugar “Steric Gate” of DNA Polymerases. Biochemistry 2011, 50, 1135-1142, 10.1021/bi101915z.
- Mary K Johnson; Jithesh Kottur; Deepak T. Nair; A polar filter in DNA polymerases prevents ribonucleotide incorporation. Nucleic Acids Research 2019, 47, 10693-10705, 10.1093/nar/gkz792.
- Andrei Chabes; Bilyana Georgieva; Vladimir Domkin; Xiaolan Zhao; Rodney Rothstein; Lars Thelander; Survival of DNA damage in yeast directly depends on increased dNTP levels allowed by relaxed feedback inhibition of ribonucleotide reductase.. Cell 2003, 112, 391-401, 10.1016/s0092-8674(03)00075-8.
- Stephanie A. Nick McElhinny; Brian E. Watts; Dinesh Kumar; Danielle L. Watt; Else-Britt Lundström; Peter M. J. Burgers; Erik Johansson; Andrei Chabes; Thomas Kunkel; Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proceedings of the National Academy of Sciences 2010, 107, 4949-4954, 10.1073/pnas.0914857107.
- Jessica S. Williams; Anders Ranegaard Clausen; Stephanie A. Nick McElhinny; Brian E. Watts; Erik Johansson; Thomas Kunkel; Proofreading of ribonucleotides inserted into DNA by yeast DNA polymerase ɛ.. DNA Repair 2012, 11, 649-56, 10.1016/j.dnarep.2012.05.004.
- Anders Ranegaard Clausen; Sufang Zhang; Peter M. Burgers; Marietta Y. Lee; Thomas Kunkel; Ribonucleotide incorporation, proofreading and bypass by human DNA polymerase δ.. DNA Repair 2012, 12, 121-7, 10.1016/j.dnarep.2012.11.006.
- Kyung Duk Koh; Sathya Balachander; Jay R. Hesselberth; Francesca Storici; Ribose-seq: global mapping of ribonucleotides embedded in genomic DNA. Nature Methods 2015, 12, 251-257, 10.1038/nmeth.3259.
- Justin L. Sparks; Hyongi Chon; Susana M. Cerritelli; Thomas Kunkel; Erik Johansson; Robert J. Crouch; Peter M. Burgers; RNase H2-Initiated Ribonucleotide Excision Repair. Molecular Cell 2012, 47, 980-6, 10.1016/j.molcel.2012.06.035.
- Stephanie A. Nick McElhinny; Dinesh Kumar; Alan B. Clark; Danielle L. Watt; Brian E. Watts; Else-Britt Lundström; Erik Johansson; Andrei Chabes; Thomas Kunkel; Genome instability due to ribonucleotide incorporation into DNA. Nature Methods 2010, 6, 774-81, 10.1038/nchembio.424.
- Martin Reijns; Björn Rabe; Rachel Rigby; Pleasantine Mill; Katy R. Astell; Laura A. Lettice; Shelagh Boyle; Andrea Leitch; Margaret Keighren; Fiona Kilanowski; et al.Paul S. DevenneyDavid SextonGraeme GrimesIan HoltRobert E. HillMartin S. TaylorKirstie A. LawsonJulia R. DorinAndrew Jackson Enzymatic Removal of Ribonucleotides from DNA Is Essential for Mammalian Genome Integrity and Development. Cell 2012, 149, 1008-22, 10.1016/j.cell.2012.04.011.
- Tomoo Ogi; Siripan Limsirichaikul; René M. Overmeer; Marcel Volker; Katsuya Takenaka; Ross Cloney; Yuka Nakazawa; Atsuko Niimi; Yoshio Miki; Nicolaas G. Jaspers; et al.Leon Hf MullendersShunichi YamashitaMaria FousteriAlan R. Lehmann Three DNA Polymerases, Recruited by Different Mechanisms, Carry Out NER Repair Synthesis in Human Cells. Molecular Cell 2010, 37, 714-727, 10.1016/j.molcel.2010.02.009.
- Alexandra Vaisman; Roger Woodgate; Ribonucleotide Discrimination by Translesion Synthesis DNA Polymerases. Critical Reviews in Biochemistry and Molecular Biology 2018, 53, 382-402, 10.1080/10409238.2018.1483889.
- Giulia Maria Nava; Lavinia Grasso; Sarah Sertic; Achille Pellicioli; Marco Muzi Falconi; Federico Lazzaro; One, No One, and One Hundred Thousand: The Many Forms of Ribonucleotides in DNA. International Journal of Molecular Sciences 2020, 21, 1706, 10.3390/ijms21051706.
- Zhiyong Mao; Michael Bozzella; Andrei Seluanov; Vera Gorbunova; DNA repair by nonhomologous end joining and homologous recombination during cell cycle in human cells.. Cell Cycle 2008, 7, 2902-6, 10.4161/cc.7.18.6679.
- Gastón Soria; Laura Belluscio; R. Kanaar; W.A. Van Cappellen; J. Essers; Vanesa Gottifredi; DNA damage induced Pol η recruitment takes place independently of the cell cycle phase. Cell Cycle 2009, 8, 3340-3348, 10.4161/cc.8.20.9836.
- Noam Diamant; Ayal Hendel; Ilan Vered; Thomas Carell; Thomas Reissner; Niels De Wind; Nicholas Geacinov; Zvi Livneh; Thomas Reißner; DNA damage bypass operates in the S and G2 phases of the cell cycle and exhibits differential mutagenicity.. Nucleic Acids Research 2011, 40, 170-80, 10.1093/nar/gkr596.
- Sarah Sertic; Antonio Mollica; Ilaria Campus; Stefania Roma; Emanuela Tumini; Andrés Aguilera; Marco Muzi Falconi; Coordinated Activity of Y Family TLS Polymerases and EXO1 Protects Non-S Phase Cells from UV-Induced Cytotoxic Lesions. Molecular Cell 2018, 70, 34-47.e4, 10.1016/j.molcel.2018.02.017.
- Ming Zhuo; Murat F. Gorgun; Ella W. Englander; Translesion Synthesis DNA Polymerase Kappa Is Indispensable for DNA Repair Synthesis in Cisplatin Exposed Dorsal Root Ganglion Neurons. Molecular Neurobiology 2017, 55, 2506-2515, 10.1007/s12035-017-0507-5.
- Susana M. Cerritelli; Robert J. Crouch; Ribonuclease H: the enzymes in eukaryotes.. The FEBS Journal 2008, 276, 1494-505, 10.1111/j.1742-4658.2009.06908.x.
- Ryo Uehara; Susana M. Cerritelli; Naushaba Hasin; Kiran Sakhuja; Mariya London; Jaime Iranzo; Hyongi Chon; Alexander Grinberg; Robert J. Crouch; Two RNase H2 Mutants with Differential rNMP Processing Activity Reveal a Threshold of Ribonucleotide Tolerance for Embryonic Development.. Cell Reports 2018, 25, 1135-1145.e5, 10.1016/j.celrep.2018.10.019.
- Yanick J. Crow; Andrea Leitch; Bruce E Hayward; Anna Garner; Rekha Parmar; Elen Griffith; Manir Ali; Colin A. Semple; Jean Aicardi; Riyana Babul-Hirji; et al.Clarisse BaumannPeter BaxterEnrico Silvio BertiniKate E ChandlerDavid ChitayatDaniel CauCatherine DéryElisa FazziCyril GoizetMary D KingJoerg KlepperDidier LacombeGiovanni LanziHermione LyallMaría Luisa Martínez-FríasMichèle MathieuCarole McKeownAnne MonierYvette OadeOliver W QuarrellChristopher D RitteyR Curtis RogersAmparo SanchisJohn B P StephensonUta TackeMarianne TillJohn L TolmiePam TomlinThomas VoitBernhard WeschkeC. Geoffrey WoodsPierre LebonDavid T BonthronChris P. PontingAndrew Jackson Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutières syndrome and mimic congenital viral brain infection. Nature Genetics 2006, 38, 910-916, 10.1038/ng1842.
- Yanick J. Crow; John H. Livingston; Neurologic Phenotypes Associated with Mutations in TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, ADAR1, and IFIH1: Aicardi–Goutières Syndrome and Beyond. Neuropediatrics 2016, 47, 355-360, 10.1055/s-0036-1592307.
- Sara Pizzi; Sarah Sertic; Simona Orcesi; Cristina Cereda; Marika Bianchi; Andrew Jackson; Federico Lazzaro; Paolo Plevani; Marco Muzi Falconi; Reduction of hRNase H2 activity in Aicardi-Goutières syndrome cells leads to replication stress and genome instability.. Human Molecular Genetics 2014, 24, 649-58, 10.1093/hmg/ddu485.
- Claudia Günther; Barbara Kind; Martin Reijns; Nicole Berndt; Manuel Martinez Bueno; Christine Wolf; Victoria Tüngler; Osvaldo Chara; Young Ae Lee; Norbert Hubner; et al.Louise BicknellSophia BlumClaudia KrugFranziska SchmidtStefanie KretschmerSarah KossKaty R. AstellGeorgia RamantaniAnja BauerfeindDavid L. MorrisDeborah S. Cunninghame GrahamRyen BubeckAndrea LeitchStuart H. RalstonElizabeth A. BlackburnManfred GahrTorsten WitteTimothy J. VyseInga MelchersElisabeth MangoldMarkus NöthenMartin AringerAnnegret KuhnKirsten LüthkeLeonore UngerAnnette BleyAlice LorenziJohn D. IsaacsDimitra AlexopoulouKarsten ConradAndreas DahlAxel RoersMarta E. Alarcon-RiquelmeAndrew JacksonMin Ae Lee-Kirsch Defective removal of ribonucleotides from DNA promotes systemic autoimmunity. Journal of Clinical Investigation 2014, 125, 413-424, 10.1172/JCI78001.
- Susana M. Cerritelli; Ella G. Frolova; Chiguang Feng; Alexander Grinberg; Paul E. Love; Robert J. Crouch; Failure to Produce Mitochondrial DNA Results in Embryonic Lethality in Rnaseh1 Null Mice. Molecular Cell 2003, 11, 807-815, 10.1016/s1097-2765(03)00088-1.
- Federico Lazzaro; Daniele Novarina; Flavio Amara; Danielle L. Watt; Jana E. Stone; Vincenzo Costanzo; Peter M. Burgers; Thomas Kunkel; Paolo Plevani; Marco Muzi Falconi; et al. RNase H and Postreplication Repair Protect Cells from Ribonucleotides Incorporated in DNA. Molecular Cell 2012, 45, 99-110, 10.1016/j.molcel.2011.12.019.
- Jessica S. Williams; Anders Ranegaard Clausen; Scott Lujan; Lisette Marjavaara; Alan B. Clark; Peter M. Burgers; Andrei Chabes; Thomas Kunkel; Evidence that processing of ribonucleotides in DNA by topoisomerase 1 is leading-strand specific. Nature Structural & Molecular Biology 2015, 22, 291-297, 10.1038/nsmb.2989.
- Katherine A. Donigan; Susana M. Cerritelli; John P. McDonald; Alexandra Vaisman; Robert J. Crouch; Roger Woodgate; Unlocking the steric gate of DNA polymerase η leads to increased genomic instability in Saccharomyces cerevisiae. DNA Repair 2015, 35, 1-12, 10.1016/j.dnarep.2015.07.002.
- Katrin Kreisel; Martin K M Engqvist; Josephine Kalm; Liam J Thompson; Martin Boström; Clara Navarrete; John P McDonald; Erik Larsson; Roger Woodgate; Anders Ranegaard Clausen; et al. DNA polymerase η contributes to genome-wide lagging strand synthesis.. Nucleic Acids Research 2019, 47, 2425-2435, 10.1093/nar/gky1291.
- Alice Meroni; Giulia Maria Nava; Eliana Bianco; Lavinia Grasso; Elena Galati; Maria Cristina Bosio; Daria Delmastro; Marco Muzi Falconi; Federico Lazzaro; RNase H activities counteract a toxic effect of Polymerase η in cells replicating with depleted dNTP pools.. Nucleic Acids Research 2019, 47, 4612-4623, 10.1093/nar/gkz165.
- Yong Hwan Jin; Rao Ayyagari; Michael Resnick; Dmitry A Gordenin; Peter M. Burgers; Okazaki Fragment Maturation in Yeast. Journal of Biological Chemistry 2002, 278, 1626-1633, 10.1074/jbc.m209803200.
- Lata Balakrishnan; Robert A. Bambara; Okazaki Fragment Metabolism. Cold Spring Harbor Perspectives in Biology 2013, 5, a010173-a010173, 10.1101/cshperspect.a010173.
- Michele Giannattasio; Dana Branzei; DNA Replication Through Strand Displacement During Lagging Strand DNA Synthesis in Saccharomyces cerevisiae. Genes 2019, 10, 167, 10.3390/genes10020167.
- Robert A. Bambara; Richard S. Murante; Leigh A. Henricksen; Enzymes and Reactions at the Eukaryotic DNA Replication Fork. Journal of Biological Chemistry 1997, 272, 4647-4650, 10.1074/jbc.272.8.4647.
- Yuan Liu; Hui-I Kao; Robert A. Bambara; FLAP E NDONUCLEASE 1: A Central Component of DNA Metabolism. Annual Review of Biochemistry 2004, 73, 589-615, 10.1146/annurev.biochem.73.012803.092453.
- Malik Kahli; Joseph S Osmundson; Rani Yeung; Duncan J. Smith; Processing of eukaryotic Okazaki fragments by redundant nucleases can be uncoupled from ongoing DNA replication in vivo.. Nucleic Acids Research 2019, 47, 1814-1822, 10.1093/nar/gky1242.
- Bochao Liu; Jiazhi Hu; Jingna Wang; Daochun Kong; Direct Visualization of RNA-DNA Primer Removal from Okazaki Fragments Provides Support for Flap Cleavage and Exonucleolytic Pathways in Eukaryotic Cells. Journal of Biological Chemistry 2017, 292, 4777-4788, 10.1074/jbc.m116.758599.
- Emily Yun-Chia Chang; Shuhe Tsai; Maria J. Aristizabal; James P. Wells; Yan Coulombe; Franciele F. Busatto; Yujia A. Chan; Arun Kumar; Yi Dan Zhu; Alan Ying-Hsu Wang; et al.Louis-Alexandre FournierPhilip HieterMichael S. KoborJean-Yves MassonPeter C. Stirling MRE11-RAD50-NBS1 promotes Fanconi Anemia R-loop suppression at transcription-replication conflicts.. Nature Communications 2019, 10, 4265-15, 10.1038/s41467-019-12271-w.
- Dmitry A Gordenin; Thomas Kunkel; Michael Resnick; Repeat expansion — all in flap?. Nature Genetics 1997, 16, 116-118, 10.1038/ng0697-116.
- Anastasiya Epshtein; Catherine J. Potenski; Hannah Klein; Increased spontaneous recombination in RNase H2-deficient cells arises from multiple contiguous rNMPs and not from single rNMP residues incorporated by DNA polymerase epsilon. Microbial Cell 2016, 3, 248-254, 10.15698/mic2016.06.506.
- J. Bradley Holmes; Gökhan Akman; Stuart R. Wood; Kiran Sakhuja; Susana M. Cerritelli; Chloe Moss; Mark R. Bowmaker; Howard T Jacobs; Robert J. Crouch; Ian Holt; et al. Primer retention owing to the absence of RNase H1 is catastrophic for mitochondrial DNA replication.. Proceedings of the National Academy of Sciences 2015, 112, 9334-9, 10.1073/pnas.1503653112.
- Sonya Vengrova; Jacob Dalgaard; RNase-sensitive DNA modification(s) initiates S. pombe mating-type switching. Genome Research 2004, 18, 794-804, 10.1101/gad.289404.
- Sonya Vengrova; Jacob Dalgaard; The wild‐type Schizosaccharomyces pombe mat1 imprint consists of two ribonucleotides. EMBO reports 2006, 7, 59-65, 10.1038/sj.embor.7400576.
- M. Thomas; R. L. White; R. W. Davis; Hybridization of RNA to double-stranded DNA: formation of R-loops.. Proceedings of the National Academy of Sciences 1976, 73, 2294-2298, 10.1073/pnas.73.7.2294.
- Kenneth Dale Westover; David A. Bushnell; Roger D. Kornberg; Structural Basis of Transcription. Cell 2004, 119, 481-489, 10.1016/j.cell.2004.10.016.
- Lamia Wahba; Lorenzo Costantino; Frederick J. Tan; Anjali Zimmer; Douglas Koshland; S1-DRIP-seq identifies high expression and polyA tracts as major contributors to R-loop formation. Genes & Development 2016, 30, 1327-1338, 10.1101/gad.280834.116.
- Paul A. Ginno; Paul C Lott; Holly C. Christensen; Ian Korf; Frederic Chedin; R-Loop Formation Is a Distinctive Characteristic of Unmethylated Human CpG Island Promoters. Molecular Cell 2012, 45, 814-25, 10.1016/j.molcel.2012.01.017.
- Maikel Castellano-Pozo; José M. Santos-Pereira; Ana G. Rondón; Sonia Barroso; Eloisa Andujar; Mónica Pérez-Alegre; Tatiana Garcia-Muse; Andrés Aguilera; R Loops Are Linked to Histone H3 S10 Phosphorylation and Chromatin Condensation. Molecular Cell 2013, 52, 583-590, 10.1016/j.molcel.2013.10.006.
- Stephan Hamperl; Karlene A. Cimprich; The contribution of co-transcriptional RNA:DNA hybrid structures to DNA damage and genome instability.. DNA Repair 2014, 19, 84-94, 10.1016/j.dnarep.2014.03.023.
- Wenjian Gan; Zhishuang Guan; Jie Liu; Ting Gui; Keng Shen; James L Manley; Xialu Li; R-loop-mediated genomic instability is caused by impairment of replication fork progression. Genes & Development 2011, 25, 2041-2056, 10.1101/gad.17010011.
- Lorenzo Costantino; Douglas Koshland; Genome-wide Map of R-Loop-Induced Damage Reveals How a Subset of R-Loops Contributes to Genomic Instability. Molecular Cell 2018, 71, 487-497.e3, 10.1016/j.molcel.2018.06.037.
- Hannah E. Mischo; Belén Gómez-González; Pawel Grzechnik; Ana G. Rondón; Wu Wei; Lars M. Steinmetz; Andrés Aguilera; Nick J. Proudfoot; Yeast Sen1 Helicase Protects the Genome from Transcription-Associated Instability. Molecular Cell 2011, 41, 21-32, 10.1016/j.molcel.2010.12.007.
- Prasun Chakraborty; Frank Grosse; Human DHX9 helicase preferentially unwinds RNA-containing displacement loops (R-loops) and G-quadruplexes. DNA Repair 2011, 10, 654-665, 10.1016/j.dnarep.2011.04.013.
- Jean-Baptiste Boulé; Virginia A. Zakian; The yeast Pif1p DNA helicase preferentially unwinds RNA DNA substrates.. Nucleic Acids Research 2007, 35, 5809-18, 10.1093/nar/gkm613.
- Tokio Kogoma; A novel Escherichia coli mutant capable of DNA replication in the absence of protein synthesis. Journal of Molecular Biology 1978, 121, 55-69, 10.1016/0022-2836(78)90262-0.
- T Kogoma; R R Maldonado; DNA polymerase I in constitutive stable DNA replication in Escherichia coli.. Journal of Bacteriology 1997, 179, 2109-2115, 10.1128/jb.179.7.2109-2115.1997.
- Richard T. Pomerantz; Michael E. O’Donnell; The replisome uses mRNA as a primer after colliding with RNA polymerase. Nature 2008, 456, 762-6, 10.1038/nature07527.
- Ruth Stuckey; Nestor Garcia Rodriguez; Andrés Aguilera; Ralf Erik Wellinger; Role for RNA:DNA hybrids in origin-independent replication priming in a eukaryotic system. Proceedings of the National Academy of Sciences 2015, 112, 5779-5784, 10.1073/pnas.1501769112.
- Nadine Puget; Kyle M. Miller; Gaëlle Legube; Non-canonical DNA/RNA structures during Transcription-Coupled Double-Strand Break Repair: Roadblocks or Bona fide repair intermediates?. DNA Repair 2019, 81, 102661, 10.1016/j.dnarep.2019.102661.
- Sonia Jimeno; Rosario- Prados- Carvajal; Pablo Huertas; The role of RNA and RNA-related proteins in the regulation of DNA double strand break repair pathway choice. DNA Repair 2019, 81, 102662, 10.1016/j.dnarep.2019.102662.
- Aldo S. Bader; Ben R. Hawley; Ania Wilczynska; Martin Bushell; The roles of RNA in DNA double-strand break repair.. British Journal of Cancer 2020, 122, 613-623, 10.1038/s41416-019-0624-1.
- John M. Pryor; Michael P. Conlin; Juan Carvajal-Garcia; Megan E. Luedeman; Adam J. Luthman; George W. Small; Dale A. Ramsden; Ribonucleotide incorporation enables repair of chromosome breaks by nonhomologous end joining. Science 2018, 361, 1126-1129, 10.1126/science.aat2477.
- Chance Meers; Havva Keskin; Francesca Storici; DNA repair by RNA: Templated, or not templated, that is the question.. DNA Repair 2016, 44, 17-21, 10.1016/j.dnarep.2016.05.002.
- Lei Li; Devon R. Germain; Ho-Yin Poon; Matthew Hildebrandt; Elizabeth A. Monckton; Darin McDonald; Michael J. Hendzel; Roseline Godbout; DEAD Box 1 Facilitates Removal of RNA and Homologous Recombination at DNA Double-Strand Breaks. Molecular and Cellular Biology 2016, 36, 2794-2810, 10.1128/MCB.00415-16.
- Corina Ohle; Rafael Tesorero; Géza Schermann; Nikolay Dobrev; Irmgard Sinning; Tamas Fischer; Transient RNA-DNA Hybrids Are Required for Efficient Double-Strand Break Repair. Cell 2016, 167, 1001-1013.e7, 10.1016/j.cell.2016.10.001.
- Julien Brustel; Zuzanna Kozik; Natalia Gromak; Velibor Savic; Steve Sweet; Large XPF-dependent deletions following misrepair of a DNA double strand break are prevented by the RNA:DNA helicase Senataxin. Scientific Reports 2018, 8, 3850, 10.1038/s41598-018-21806-y.
- Sarah Cohen; Nadine Puget; Yea-Lih Lin; Thomas Clouaire; Marion Aguirrebengoa; Vincent Rocher; Philippe Pasero; Yvan Canitrot; Gaëlle Legube; Senataxin resolves RNA:DNA hybrids forming at DNA double-strand breaks to prevent translocations.. Nature Communications 2018, 9, 533, 10.1038/s41467-018-02894-w.
- Giuseppina D’Alessandro; Donna R Whelan; Sean Michael Howard; Valerio Vitelli; Xavier Renaudin; Marek Adamowicz; Fabio Iannelli; Corey Winston Jones-Weinert; Miyoung Lee; Valentina Matti; et al.Wei Ting C. LeeMichael John MortenAshok Raraakrishnan VenkitaramanPetr CejkaEli RothenbergFabrizio Daddadifagagna BRCA2 controls DNA:RNA hybrid level at DSBs by mediating RNase H2 recruitment. Nature Communications 2018, 9, 5376, 10.1038/s41467-018-07799-2.
- Wei-Ting Lu; Ben R. Hawley; George Skalka; Robert A. Baldock; Ewan Smith; Aldo S. Bader; Michal Malewicz; Felicity Z. Watts; Ania Wilczynska; Martin Bushell; et al. Drosha drives the formation of DNA:RNA hybrids around DNA break sites to facilitate DNA repair. Nature Communications 2018, 9, 532, 10.1038/s41467-018-02893-x.
- Takaaki Yasuhara; Reona Kato; Yoshihiko Hagiwara; Bunsyo Shiotani; Motohiro Yamauchi; Shinichiro Nakada; Atsushi Shibata; Kiyoshi Miyagawa; Human Rad52 Promotes XPG-Mediated R-loop Processing to Initiate Transcription-Associated Homologous Recombination Repair. Cell 2018, 175, 558-570.e11, 10.1016/j.cell.2018.08.056.
- Kaspar Burger; Margarita Schlackow; Monika Gullerova; Tyrosine kinase c-Abl couples RNA polymerase II transcription to DNA double-strand breaks.. Nucleic Acids Research 2019, 47, 3467-3484, 10.1093/nar/gkz024.
- Luigi Alfano; Antonella Caporaso; Angela Altieri; Milena Dell'aquila; Claudia Landi; Luca Bini; Francesca Pentimalli; Antonio Giordano; Depletion of the RNA binding protein HNRNPD impairs homologous recombination by inhibiting DNA-end resection and inducing R-loop accumulation.. Nucleic Acids Research 2019, 47, 4068-4085, 10.1093/nar/gkz076.
- Judit Domingo Prim; Martin Endara-Coll; Franziska Bonath; Sonia Jimeno; Rosario Prados-Carvajal; Marc R Friedländer; Pablo Huertas; Neus Visa; EXOSC10 is required for RPA assembly and controlled DNA end resection at DNA double-strand breaks.. Nature Communications 2019, 10, 2135, 10.1038/s41467-019-10153-9.
- Lorraine Symington; Mechanism and regulation of DNA end resection in eukaryotes.. Critical Reviews in Biochemistry and Molecular Biology 2016, 51, 195-212, 10.3109/10409238.2016.1172552.
- Federica Marini; Chetan C. Rawal; Giordano Liberi; Achille Pellicioli; Regulation of DNA Double Strand Breaks Processing: Focus on Barriers.. Frontiers in Molecular Biosciences 2019, 6, 55, 10.3389/fmolb.2019.00055.
- Hyongi Chon; Justin L. Sparks; Monika Rychlik; Marcin Nowotny; Peter M. Burgers; Robert J. Crouch; Susana M. Cerritelli; RNase H2 roles in genome integrity revealed by unlinking its activities. Nucleic Acids Research 2013, 41, 3130-3143, 10.1093/nar/gkt027.
- Susana M. Cerritelli; Robert J. Crouch; RNase H2-RED carpets the path to eukaryotic RNase H2 functions. DNA Repair 2019, 84, 102736, 10.1016/j.dnarep.2019.102736.
- Maria Falkenberg; Mitochondrial DNA replication in mammalian cells: overview of the pathway. Essays in Biochemistry 2018, 62, 287-296, 10.1042/ebc20170100.
- Takehiro Yasukawa; Ngchon Kang; An overview of mammalian mitochondrial DNA replication mechanisms. The Journal of Biochemistry 2018, 164, 183-193, 10.1093/jb/mvy058.
- Sjoerd Wanrooij; Javier Miralles Fusté; Géraldine Farge; Yonghong Shi; Claes M. Gustafsson; Maria Falkenberg; Human mitochondrial RNA polymerase primes lagging-strand DNA synthesis in vitro. Proceedings of the National Academy of Sciences 2008, 105, 11122-11127, 10.1073/pnas.0805399105.
- Paulina Wanrooij; Jay P. Uhler; Yonghong Shi; Fredrik Westerlund; Maria Falkenberg; Claes M. Gustafsson; A hybrid G-quadruplex structure formed between RNA and DNA explains the extraordinary stability of the mitochondrial R-loop.. Nucleic Acids Research 2012, 40, 10334-44, 10.1093/nar/gks802.
- Takehiro Yasukawa; Aurelio Reyes; Tricia J Cluett; Mingyao Yang; Mark Bowmaker; Howard T Jacobs; Ian Holt; Replication of vertebrate mitochondrial DNA entails transient ribonucleotide incorporation throughout the lagging strand. The EMBO Journal 2006, 25, 5358-5371, 10.1038/sj.emboj.7601392.
- Aurelio Reyes; Laura Melchionda; Alessia Nasca; Franco Carrara; Eleonora Lamantea; Alice Zanolini; Costanza Lamperti; Mingyan Fang; Jianguo Zhang; Dario Ronchi; et al.Sara BonatoGigliola FagiolariMaurizio MoggioDaniele GhezziMassimo Zeviani RNASEH1 Mutations Impair mtDNA Replication and Cause Adult-Onset Mitochondrial Encephalomyopathy. Expanding the Spectrum of BAF-Related Disorders: De Novo Variants in SMARCC2 Cause a Syndrome with Intellectual Disability and Developmental Delay 2015, 97, 186-193, 10.1016/j.ajhg.2015.05.013.
- Stewart A. Morley; Niaz Ahmad; Brent Nielsen; Plant Organelle Genome Replication.. Plants 2019, 8, 358, 10.3390/plants8100358.
- Zhuo Yang; Quancan Hou; Lingling Cheng; Wei Xu; Yantao Hong; Shuai Li; Qianwen Sun; RNase H1 Cooperates with DNA Gyrases to Restrict R-Loops and Maintain Genome Integrity in Arabidopsis Chloroplasts. The Plant Cell 2017, 29, 2478-2497, 10.1105/tpc.17.00305.
- Jan Kuciński; Aleksandra Kmera; M. Jordan Rowley; Pragya Khurana; Marcin Nowotny; Andrzej T Wierzbicki; Functional characterization of RNase H1 proteins in Arabidopsis thaliana. bioRxiv 2019, -, 662080, 10.1101/662080.