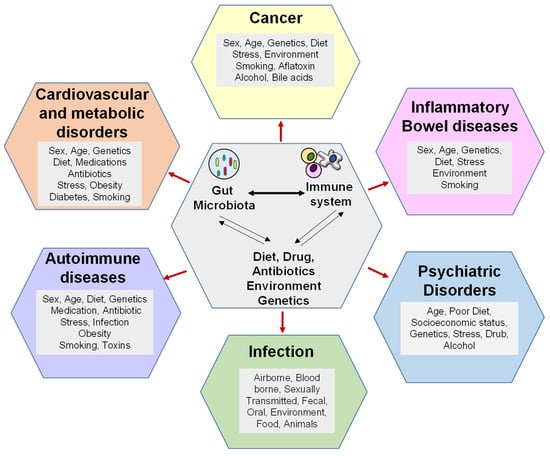
Gut microbes and their metabolites are actively involved in the development and regulation of host immunity, which can influence disease susceptibility. The role of the microbiome as a protective force is supported by research indicating that immature microbiomes of neonates are more susceptible to invasion by pathobionts. During early years of development, exposure to various microbes shapes the immune system for a lifetime. The gut microbiome has a wide range of metabolic activities, including metabolizing lipids, carbohydrates, and proteins, and participates in maintaining host homeostasis. Therefore, disruption of gut microbiome can lead to conditions as severe as cancer.
- gut microbiota dysbiosis
- innate immune system
- adaptive immune system
1. Gut Microbiota Dysbiosis and Immune Dysregulation
| Diseases | Reference | Findings |
|---|---|---|
| Gastrointestinal Infections | Singer et al., 2019 [10] | Provide resistance against colonization and invasion by pathobiont. |
| Tovaglieri et al., 2019 [11] | Human gut microbiome metabolites induce expression of flagellin (a bacterial protein) increases EHEC motility and epithelial injury. | |
| IBD | Lee and chang, 2021 [12] | Gut microbiota dysbiosis of IBD patients is consistently marked by an overgrowth in Proteobacteria. |
| Furusawa et al., 2013 [13] | SCFA confers protection against IBD by maintaining gut barrier integrity, promoting Treg cell differentiation, and inhibiting histone deacetylases. | |
| Colorectal carcinoma |
Sepich-Poore et al., 2021 [14] | Generation of genotoxin such as Bacteroides fragilis toxin (Bft), cytolethal distending toxin (CDT), and colibactin. |
| Hale et al., 2017 [15] | Bacterial-derived secondary bile acids and hydrogen sulfide promote proinflammatory milieu that increases CRC risk. | |
| Yeoh et al., 2020 [16] | Bacteria such as F. nucleatum can adhere to colon tumors and aggravate tumorigenesis. | |
| Hepatocellular carcinoma | Lin et al., 1995 [17] | Systemic translocation of LPS promotes chronic liver injury and predisposes to HCC. |
| Singh et al., 2018 [18] | Excess butyrate production promotes HCC progression. | |
| Yoshimoto et al., 2013 [19] | Secondary bile acids promote carcinogenesis and impede anti-tumor immunosurveillance in the liver. | |
| Cardiometabolic disease | Cani et al., 2007 [20] Guasch-Ferré et al., 2017 [21] Millard et al., 2018 [22] |
LPS and other microbial ligands drive low-grade chronic inflammation and predispose to CVD. |
| Bacterial trimethylamine and its conversion to trimethylamine-N-oxide in the liver increases the risk of coronary artery disease, metabolic syndrome, stroke, and vascular inflammation. | ||
| Rheumatoid Arthritis |
Scher et al., 2013 [23] | Prevotella spp. Abundance is positively associated with new-onset rheumatoid arthritis. |
| Allergic Diseases |
Fazlollahi et al., 2018 [24] Bunyavanich et al., 2016 [25] |
Gut microbiota dysbiosis increases risk for allergic disease, e.g., food allergy and asthma. |
2. Gastrointestinal Infections
3. Inflammatory Bowel Diseases
4. Colorectal Carcinoma (CRC)
5. Hepatocellular Carcinoma (HCC)
6. Cardiovascular Disease
7. Diabetes
8. Hypertension
9. Rheumatoid Arthritis
10. Allergic Diseases
11. Psychiatric Disorders: The Gut–Brain Axis
This entry is adapted from the peer-reviewed paper 10.3390/biomedicines11020294
References
- Mager, L.F.; Burkhard, R.; Pett, N.; Cooke, N.C.A.; Brown, K.; Ramay, H.; Paik, S.; Stagg, J.; Groves, R.A.; Gallo, M.; et al. Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy. Science 2020, 369, 1481–1489.
- Ahmadi, S.; Razazan, A.; Nagpal, R.; Jain, S.; Wang, B.; Mishra, S.P.; Wang, S.; Justice, J.; Ding, J.; McClain, D.A.; et al. Metformin Reduces Aging-Related Leaky Gut and Improves Cognitive Function by Beneficially Modulating Gut Microbiome/Goblet Cell/Mucin Axis. J. Gerontol. A Biol. Sci. Med. Sci. 2020, 75, e9–e21.
- Stevens, B.R.; Goel, R.; Seungbum, K.; Richards, E.M.; Holbert, R.C.; Pepine, C.J.; Raizada, M.K. Increased human intestinal barrier permeability plasma biomarkers zonulin and FABP2 correlated with plasma LPS and altered gut microbiome in anxiety or depression. Gut 2018, 67, 1555–1557.
- Thevaranjan, N.; Puchta, A.; Schulz, C.; Naidoo, A.; Szamosi, J.C.; Verschoor, C.P.; Loukov, D.; Schenck, L.P.; Jury, J.; Foley, K.P.; et al. Age-Associated Microbial Dysbiosis Promotes Intestinal Permeability, Systemic Inflammation, and Macrophage Dysfunction. Cell Host Microbe 2017, 21, 455–466.
- Hamer, H.M.; De Preter, V.; Windey, K.; Verbeke, K. Functional analysis of colonic bacterial metabolism: Relevant to health? Am. J. Physiol. Gastrointest. Liver Physiol. 2012, 302, G1–G9.
- McCall, I.C.; Betanzos, A.; Weber, D.A.; Nava, P.; Miller, G.W.; Parkos, C.A. Effects of phenol on barrier function of a human intestinal epithelial cell line correlate with altered tight junction protein localization. Toxicol. Appl. Pharmacol. 2009, 241, 61–70.
- Cox, L.M.; Yamanishi, S.; Sohn, J.; Alekseyenko, A.V.; Leung, J.M.; Cho, I.; Kim, S.G.; Li, H.; Gao, Z.; Mahana, D.; et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell 2014, 158, 705–721.
- Jacobs, M.C.; Haak, B.W.; Hugenholtz, F.; Wiersinga, W.J. Gut microbiota and host defense in critical illness. Curr. Opin. Crit. Care 2017, 23, 257–263.
- Van der Poll, T.; van de Veerdonk, F.L.; Scicluna, B.P.; Netea, M.G. The immunopathology of sepsis and potential therapeutic targets. Nat. Rev. Immunol. 2017, 17, 407–420.
- Singer, J.R.; Blosser, E.G.; Zindl, C.L.; Silberger, D.J.; Conlan, S.; Laufer, V.A.; DiToro, D.; Deming, C.; Kumar, R.; Morrow, C.D.; et al. Preventing dysbiosis of the neonatal mouse intestinal microbiome protects against late-onset sepsis. Nat. Med. 2019, 25, 1772–1782.
- Tovaglieri, A.; Sontheimer-Phelps, A.; Geirnaert, A.; Prantil-Baun, R.; Camacho, D.M.; Chou, D.B.; Jalili-Firoozinezhad, S.; de Wouters, T.; Kasendra, M.; Super, M.; et al. Species-specific enhancement of enterohemorrhagic E. coli pathogenesis mediated by microbiome metabolites. Microbiome 2019, 7, 43.
- Lee, M.; Chang, E.B. Inflammatory Bowel Diseases (IBD) and the Microbiome-Searching the Crime Scene for Clues. Gastroenterology 2021, 160, 524–537.
- Furusawa, Y.; Obata, Y.; Fukuda, S.; Endo, T.A.; Nakato, G.; Takahashi, D.; Nakanishi, Y.; Uetake, C.; Kato, K.; Kato, T.; et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013, 504, 446–450.
- Sepich-Poore, G.D.; Zitvogel, L.; Straussman, R.; Hasty, J.; Wargo, J.A.; Knight, R. The microbiome and human cancer. Science 2021, 371, eabc4552.
- Hale, V.L.; Chen, J.; Johnson, S.; Harrington, S.C.; Yab, T.C.; Smyrk, T.C.; Nelson, H.; Boardman, L.A.; Druliner, B.R.; Levin, T.R.; et al. Shifts in the Fecal Microbiota Associated with Adenomatous Polyps. Cancer Epidemiol. Biomark. Prev. 2017, 26, 85–94.
- Yeoh, Y.K.; Chen, Z.; Wong, M.C.S.; Hui, M.; Yu, J.; Ng, S.C.; Sung, J.J.Y.; Chan, F.K.L.; Chan, P.K.S. Southern Chinese populations harbour non-nucleatum Fusobacteria possessing homologues of the colorectal cancer-associated FadA virulence factor. Gut 2020, 69, 1998–2007.
- Lin, R.S.; Lee, F.Y.; Lee, S.D.; Tsai, Y.T.; Lin, H.C.; Lu, R.H.; Hsu, W.C.; Huang, C.C.; Wang, S.S.; Lo, K.J. Endotoxemia in patients with chronic liver diseases: Relationship to severity of liver diseases, presence of esophageal varices, and hyperdynamic circulation. J. Hepatol 1995, 22, 165–172.
- Singh, V.; Yeoh, B.S.; Chassaing, B.; Xiao, X.; Saha, P.; Aguilera Olvera, R.; Lapek, J.D., Jr.; Zhang, L.; Wang, W.B.; Hao, S.; et al. Dysregulated Microbial Fermentation of Soluble Fiber Induces Cholestatic Liver Cancer. Cell 2018, 175, 679–694.
- Yoshimoto, S.; Loo, T.M.; Atarashi, K.; Kanda, H.; Sato, S.; Oyadomari, S.; Iwakura, Y.; Oshima, K.; Morita, H.; Hattori, M.; et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature 2013, 499, 97–101.
- Cani, P.D.; Amar, J.; Iglesias, M.A.; Poggi, M.; Knauf, C.; Bastelica, D.; Neyrinck, A.M.; Fava, F.; Tuohy, K.M.; Chabo, C.; et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 2007, 56, 1761–1772.
- Guasch-Ferre, M.; Hu, F.B.; Ruiz-Canela, M.; Bullo, M.; Toledo, E.; Wang, D.D.; Corella, D.; Gomez-Gracia, E.; Fiol, M.; Estruch, R.; et al. Plasma Metabolites from Choline Pathway and Risk of Cardiovascular Disease in the PREDIMED (Prevention with Mediterranean Diet) Study. J. Am. Heart Assoc. 2017, 6, e006524.
- Millard, H.R.; Musani, S.K.; Dibaba, D.T.; Talegawkar, S.A.; Taylor, H.A.; Tucker, K.L.; Bidulescu, A. Dietary choline and betaine; associations with subclinical markers of cardiovascular disease risk and incidence of CVD, coronary heart disease and stroke: The Jackson Heart Study. Eur. J. Nutr. 2018, 57, 51–60.
- Scher, J.U.; Sczesnak, A.; Longman, R.S.; Segata, N.; Ubeda, C.; Bielski, C.; Rostron, T.; Cerundolo, V.; Pamer, E.G.; Abramson, S.B.; et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife 2013, 2, e01202.
- Fazlollahi, M.; Chun, Y.; Grishin, A.; Wood, R.A.; Burks, A.W.; Dawson, P.; Jones, S.M.; Leung, D.Y.M.; Sampson, H.A.; Sicherer, S.H.; et al. Early-life gut microbiome and egg allergy. Allergy 2018, 73, 1515–1524.
- Bunyavanich, S.; Shen, N.; Grishin, A.; Wood, R.; Burks, W.; Dawson, P.; Jones, S.M.; Leung, D.Y.M.; Sampson, H.; Sicherer, S.; et al. Early-life gut microbiome composition and milk allergy resolution. J. Allergy Clin. Immunol. 2016, 138, 1122–1130.
- Osbelt, L.; Wende, M.; Almasi, E.; Derksen, E.; Muthukumarasamy, U.; Lesker, T.R.; Galvez, E.J.C.; Pils, M.C.; Schalk, E.; Chhatwal, P.; et al. Klebsiella oxytoca causes colonization resistance against multidrug-resistant K. pneumoniae in the gut via cooperative carbohydrate competition. Cell Host Microbe 2021, 29, 1663–1679.
- Osbelt, L.; Thiemann, S.; Smit, N.; Lesker, T.R.; Schroter, M.; Galvez, E.J.C.; Schmidt-Hohagen, K.; Pils, M.C.; Muhlen, S.; Dersch, P.; et al. Variations in microbiota composition of laboratory mice influence Citrobacter rodentium infection via variable short-chain fatty acid production. PLoS Pathog. 2020, 16, e1008448.
- Oliveira, R.A.; Ng, K.M.; Correia, M.B.; Cabral, V.; Shi, H.; Sonnenburg, J.L.; Huang, K.C.; Xavier, K.B. Klebsiella michiganensis transmission enhances resistance to Enterobacteriaceae gut invasion by nutrition competition. Nat. Microbiol. 2020, 5, 630–641.
- Ducarmon, Q.R.; Zwittink, R.D.; Hornung, B.V.H.; van Schaik, W.; Young, V.B.; Kuijper, E.J. Gut Microbiota and Colonization Resistance against Bacterial Enteric Infection. Microbiol. Mol. Biol. Rev. 2019, 83, e00007-19.
- Jacobson, A.; Lam, L.; Rajendram, M.; Tamburini, F.; Honeycutt, J.; Pham, T.; Van Treuren, W.; Pruss, K.; Stabler, S.R.; Lugo, K.; et al. A Gut Commensal-Produced Metabolite Mediates Colonization Resistance to Salmonella Infection. Cell Host Microbe 2018, 24, 296–307.
- Izquierdo, M.; Lopez, J.; Gallardo, P.; Vidal, R.M.; Ossa, J.C.; Farfan, M.J. Bacteria from gut microbiota associated with diarrheal infections in children promote virulence of Shiga toxin-producing and enteroaggregative Escherichia coli pathotypes. Front. Cell. Infect. Microbiol. 2022, 12, 867205.
- Lesniak, N.A.; Schubert, A.M.; Flynn, K.J.; Leslie, J.L.; Sinani, H.; Bergin, I.L.; Young, V.B.; Schloss, P.D. The Gut Bacterial Community Potentiates Clostridioides difficile Infection Severity. mBio 2022, 13, e0118322.
- Thiemann, S.; Smit, N.; Roy, U.; Lesker, T.R.; Galvez, E.J.C.; Helmecke, J.; Basic, M.; Bleich, A.; Goodman, A.L.; Kalinke, U.; et al. Enhancement of IFNgamma Production by Distinct Commensals Ameliorates Salmonella-Induced Disease. Cell Host Microbe 2017, 21, 682–694.e685.
- Caballero, S.; Kim, S.; Carter, R.A.; Leiner, I.M.; Susac, B.; Miller, L.; Kim, G.J.; Ling, L.; Pamer, E.G. Cooperating Commensals Restore Colonization Resistance to Vancomycin-Resistant Enterococcus faecium. Cell Host Microbe 2017, 21, 592–602.e594.
- Bibbo, S.; Lopetuso, L.R.; Ianiro, G.; Di Rienzo, T.; Gasbarrini, A.; Cammarota, G. Role of microbiota and innate immunity in recurrent Clostridium difficile infection. J. Immunol. Res. 2014, 2014, 462740.
- Warny, M.; Keates, A.C.; Keates, S.; Castagliuolo, I.; Zacks, J.K.; Aboudola, S.; Qamar, A.; Pothoulakis, C.; LaMont, J.T.; Kelly, C.P. p38 MAP kinase activation by Clostridium difficile toxin A mediates monocyte necrosis, IL-8 production, and enteritis. J. Clin. Investig. 2000, 105, 1147–1156.
- Ishida, Y.; Maegawa, T.; Kondo, T.; Kimura, A.; Iwakura, Y.; Nakamura, S.; Mukaida, N. Essential involvement of IFN-gamma in Clostridium difficile toxin A-induced enteritis. J. Immunol. 2004, 172, 3018–3025.
- Theriot, C.M.; Koenigsknecht, M.J.; Carlson, P.E., Jr.; Hatton, G.E.; Nelson, A.M.; Li, B.; Huffnagle, G.B.; Li, J.Z.; Young, V.B. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat. Commun. 2014, 5, 3114.
- Rea, M.C.; Dobson, A.; O′Sullivan, O.; Crispie, F.; Fouhy, F.; Cotter, P.D.; Shanahan, F.; Kiely, B.; Hill, C.; Ross, R.P. Effect of broad- and narrow-spectrum antimicrobials on Clostridium difficile and microbial diversity in a model of the distal colon. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. 1), 4639–4644.
- Buffie, C.G.; Jarchum, I.; Equinda, M.; Lipuma, L.; Gobourne, A.; Viale, A.; Ubeda, C.; Xavier, J.; Pamer, E.G. Profound alterations of intestinal microbiota following a single dose of clindamycin results in sustained susceptibility to Clostridium difficile-induced colitis. Infect. Immun. 2012, 80, 62–73.
- McDonald, L.C. Effects of short- and long-course antibiotics on the lower intestinal microbiome as they relate to traveller′s diarrhea. J. Travel Med. 2017, 24, S35–S38.
- Wang, S.; Deng, W.; Li, F.; Chen, Y.E.; Wang, P.U. Blockade of T helper 17 cell function ameliorates recurrent Clostridioides difficile infection in mice. Acta Biochim. Biophys. Sin. 2021, 53, 1290–1299.
- Frisbee, A.L.; Saleh, M.M.; Young, M.K.; Leslie, J.L.; Simpson, M.E.; Abhyankar, M.M.; Cowardin, C.A.; Ma, J.Z.; Pramoonjago, P.; Turner, S.D.; et al. IL-33 drives group 2 innate lymphoid cell-mediated protection during Clostridium difficile infection. Nat. Commun. 2019, 10, 2712.
- Wu, Z.; Xu, Q.; Wang, Q.; Chen, Y.; Lv, L.; Zheng, B.; Yan, R.; Jiang, H.; Shen, J.; Wang, S.; et al. The impact of dietary fibers on Clostridioides difficile infection in a mouse model. Front. Cell. Infect. Microbiol. 2022, 12, 1028267.
- Hayashi, A.; Nagao-Kitamoto, H.; Kitamoto, S.; Kim, C.H.; Kamada, N. The Butyrate-Producing Bacterium Clostridium butyricum Suppresses Clostridioides difficile Infection via Neutrophil- and Antimicrobial Cytokine-Dependent but GPR43/109a-Independent Mechanisms. J. Immunol. 2021, 206, 1576–1585.
- Green, J.E.; Davis, J.A.; Berk, M.; Hair, C.; Loughman, A.; Castle, D.; Athan, E.; Nierenberg, A.A.; Cryan, J.F.; Jacka, F.; et al. Efficacy and safety of fecal microbiota transplantation for the treatment of diseases other than Clostridium difficile infection: A systematic review and meta-analysis. Gut Microbes 2020, 12, 1854640.
- Petrof, E.O.; Gloor, G.B.; Vanner, S.J.; Weese, S.J.; Carter, D.; Daigneault, M.C.; Brown, E.M.; Schroeter, K.; Allen-Vercoe, E. Stool substitute transplant therapy for the eradication of Clostridium difficile infection: ′RePOOPulating′ the gut. Microbiome 2013, 1, 3.
- Martz, S.L.; McDonald, J.A.; Sun, J.; Zhang, Y.G.; Gloor, G.B.; Noordhof, C.; He, S.M.; Gerbaba, T.K.; Blennerhassett, M.; Hurlbut, D.J.; et al. Administration of defined microbiota is protective in a murine Salmonella infection model. Sci. Rep. 2015, 5, 16094.
- Barbara, G.; Barbaro, M.R.; Fuschi, D.; Palombo, M.; Falangone, F.; Cremon, C.; Marasco, G.; Stanghellini, V. Inflammatory and Microbiota-Related Regulation of the Intestinal Epithelial Barrier. Front. Nutr. 2021, 8, 718356.
- Chassaing, B.; Darfeuille-Michaud, A. The commensal microbiota and enteropathogens in the pathogenesis of inflammatory bowel diseases. Gastroenterology 2011, 140, 1720–1728.
- Swidsinski, A.; Ladhoff, A.; Pernthaler, A.; Swidsinski, S.; Loening-Baucke, V.; Ortner, M.; Weber, J.; Hoffmann, U.; Schreiber, S.; Dietel, M.; et al. Mucosal flora in inflammatory bowel disease. Gastroenterology 2002, 122, 44–54.
- Schwartz, S.M.; Kemnitz, J.W. Age- and gender-related changes in body size, adiposity, and endocrine and metabolic parameters in free-ranging rhesus macaques. Am. J. Phys. Anthropol. 1992, 89, 109–121.
- Gevers, D.; Kugathasan, S.; Denson, L.A.; Vazquez-Baeza, Y.; Van Treuren, W.; Ren, B.; Schwager, E.; Knights, D.; Song, S.J.; Yassour, M.; et al. The treatment-naive microbiome in new-onset Crohn′s disease. Cell Host Microbe 2014, 15, 382–392.
- Machiels, K.; Joossens, M.; Sabino, J.; De Preter, V.; Arijs, I.; Eeckhaut, V.; Ballet, V.; Claes, K.; Van Immerseel, F.; Verbeke, K.; et al. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut 2014, 63, 1275–1283.
- Devkota, S.; Wang, Y.; Musch, M.W.; Leone, V.; Fehlner-Peach, H.; Nadimpalli, A.; Antonopoulos, D.A.; Jabri, B.; Chang, E.B. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature 2012, 487, 104–108.
- Shapiro, J.M.; de Zoete, M.R.; Palm, N.W.; Laenen, Y.; Bright, R.; Mallette, M.; Bu, K.; Bielecka, A.A.; Xu, F.; Hurtado-Lorenzo, A.; et al. Immunoglobulin A Targets a Unique Subset of the Microbiota in Inflammatory Bowel Disease. Cell Host Microbe 2021, 29, 83–93.
- Palm, N.W.; de Zoete, M.R.; Cullen, T.W.; Barry, N.A.; Stefanowski, J.; Hao, L.; Degnan, P.H.; Hu, J.; Peter, I.; Zhang, W.; et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 2014, 158, 1000–1010.
- Schaubeck, M.; Clavel, T.; Calasan, J.; Lagkouvardos, I.; Haange, S.B.; Jehmlich, N.; Basic, M.; Dupont, A.; Hornef, M.; von Bergen, M.; et al. Dysbiotic gut microbiota causes transmissible Crohn′s disease-like ileitis independent of failure in antimicrobial defence. Gut 2016, 65, 225–237.
- Torres, J.; Hu, J.; Seki, A.; Eisele, C.; Nair, N.; Huang, R.; Tarassishin, L.; Jharap, B.; Cote-Daigneault, J.; Mao, Q.; et al. Infants born to mothers with IBD present with altered gut microbiome that transfers abnormalities of the adaptive immune system to germ-free mice. Gut 2020, 69, 42–51.
- Franzosa, E.A.; Sirota-Madi, A.; Avila-Pacheco, J.; Fornelos, N.; Haiser, H.J.; Reinker, S.; Vatanen, T.; Hall, A.B.; Mallick, H.; McIver, L.J.; et al. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat. Microbiol. 2019, 4, 293–305.
- Huang, C.; Mei, Q.; Lou, L.; Huang, Z.; Fu, Y.; Fan, J.; Wang, J.; Yin, N.; Zheng, Y.; Lu, Y.; et al. Ulcerative Colitis in Response to Fecal Microbiota Transplantation via Modulation of Gut Microbiota and Th17/Treg Cell Balance. Cells 2022, 11, 1851.
- Facchin, S.; Vitulo, N.; Calgaro, M.; Buda, A.; Romualdi, C.; Pohl, D.; Perini, B.; Lorenzon, G.; Marinelli, C.; D′Inca, R.; et al. Microbiota changes induced by microencapsulated sodium butyrate in patients with inflammatory bowel disease. Neurogastroenterol. Motil. 2020, 32, e13914.
- Li, G.; Lin, J.; Zhang, C.; Gao, H.; Lu, H.; Gao, X.; Zhu, R.; Li, Z.; Li, M.; Liu, Z. Microbiota metabolite butyrate constrains neutrophil functions and ameliorates mucosal inflammation in inflammatory bowel disease. Gut Microbes 2021, 13, 1968257.
- Hartog, A.; Belle, F.N.; Bastiaans, J.; de Graaff, P.; Garssen, J.; Harthoorn, L.F.; Vos, A.P. A potential role for regulatory T-cells in the amelioration of DSS induced colitis by dietary non-digestible polysaccharides. J. Nutr. Biochem. 2015, 26, 227–233.
- Singh, V.; Yeoh, B.S.; Walker, R.E.; Xiao, X.; Saha, P.; Golonka, R.M.; Cai, J.; Bretin, A.C.A.; Cheng, X.; Liu, Q.; et al. Microbiota fermentation-NLRP3 axis shapes the impact of dietary fibres on intestinal inflammation. Gut 2019, 68, 1801–1812.
- Zhu, Y.; Xu, Y.; Wang, X.; Rao, L.; Yan, X.; Gao, R.; Shen, T.; Zhou, Y.; Kong, C.; Zhou, L. Probiotic Cocktail Alleviates Intestinal Inflammation Through Improving Gut Microbiota and Metabolites in Colitis Mice. Front. Cell. Infect. Microbiol. 2022, 12, 886061.
- Xu, M.; Shen, Y.; Cen, M.; Zhu, Y.; Cheng, F.; Tang, L.; Zheng, X.; Kim, J.J.; Dai, N.; Hu, W. Modulation of the Gut Microbiota-farnesoid X Receptor Axis Improves Deoxycholic Acid-induced Intestinal Inflammation in Mice. J. Crohn’s Colitis 2021, 15, 1197–1210.
- Xu, M.; Cen, M.; Shen, Y.; Zhu, Y.; Cheng, F.; Tang, L.; Hu, W.; Dai, N. Deoxycholic Acid-Induced Gut Dysbiosis Disrupts Bile Acid Enterohepatic Circulation and Promotes Intestinal Inflammation. Dig. Dis. Sci. 2021, 66, 568–576.
- Liu, T.C.; Kern, J.T.; Jain, U.; Sonnek, N.M.; Xiong, S.; Simpson, K.F.; VanDussen, K.L.; Winkler, E.S.; Haritunians, T.; Malique, A.; et al. Western diet induces Paneth cell defects through microbiome alterations and farnesoid X receptor and type I interferon activation. Cell Host Microbe 2021, 29, 988–1001.
- Liu, Y.; Xu, J.; Ren, X.; Zhang, Y.; Ke, Z.; Zhou, J.; Wang, Y.; Zhang, Y.; Liu, Y. Cholecystectomy-induced secondary bile acids accumulation ameliorates colitis through inhibiting monocyte/macrophage recruitment. Gut Microbes 2022, 14, 2107387.
- Van den Bossche, L.; Hindryckx, P.; Devisscher, L.; Devriese, S.; Van Welden, S.; Holvoet, T.; Vilchez-Vargas, R.; Vital, M.; Pieper, D.H.; Vanden Bussche, J.; et al. Ursodeoxycholic Acid and Its Taurine- or Glycine-Conjugated Species Reduce Colitogenic Dysbiosis and Equally Suppress Experimental Colitis in Mice. Appl. Environ. Microbiol. 2017, 83, e02766-16.
- Duboc, H.; Rajca, S.; Rainteau, D.; Benarous, D.; Maubert, M.A.; Quervain, E.; Thomas, G.; Barbu, V.; Humbert, L.; Despras, G.; et al. Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut 2013, 62, 531–539.
- Moltzau Anderson, J.; Lipinski, S.; Sommer, F.; Pan, W.H.; Boulard, O.; Rehman, A.; Falk-Paulsen, M.; Stengel, S.T.; Aden, K.; Hasler, R.; et al. NOD2 Influences Trajectories of Intestinal Microbiota Recovery After Antibiotic Perturbation. Cell. Mol. Gastroenterol. Hepatol. 2020, 10, 365–389.
- Danne, C.; Michaudel, C.; Skerniskyte, J.; Planchais, J.; Magniez, A.; Agus, A.; Michel, M.L.; Lamas, B.; Da Costa, G.; Spatz, M.; et al. CARD9 in neutrophils protects from colitis and controls mitochondrial metabolism and cell survival. Gut 2022.
- Lavoie, S.; Conway, K.L.; Lassen, K.G.; Jijon, H.B.; Pan, H.; Chun, E.; Michaud, M.; Lang, J.K.; Gallini Comeau, C.A.; Dreyfuss, J.M.; et al. The Crohn′s disease polymorphism, ATG16L1 T300A, alters the gut microbiota and enhances the local Th1/Th17 response. Elife 2019, 8, e39982.
- Aden, K.; Tran, F.; Ito, G.; Sheibani-Tezerji, R.; Lipinski, S.; Kuiper, J.W.; Tschurtschenthaler, M.; Saveljeva, S.; Bhattacharyya, J.; Hasler, R.; et al. ATG16L1 orchestrates interleukin-22 signaling in the intestinal epithelium via cGAS-STING. J. Exp. Med. 2018, 215, 2868–2886.
- Zhou, C.B.; Zhou, Y.L.; Fang, J.Y. Gut Microbiota in Cancer Immune Response and Immunotherapy. Trends Cancer 2021, 7, 647–660.
- Xu, C.; Fan, L.; Lin, Y.; Shen, W.; Qi, Y.; Zhang, Y.; Chen, Z.; Wang, L.; Long, Y.; Hou, T.; et al. Fusobacterium nucleatum promotes colorectal cancer metastasis through miR-1322/CCL20 axis and M2 polarization. Gut Microbes 2021, 13, 1980347.
- Hussan, H.; Clinton, S.K.; Roberts, K.; Bailey, M.T. Fusobacterium′s link to colorectal neoplasia sequenced: A systematic review and future insights. World J. Gastroenterol. 2017, 23, 8626–8650.
- Yang, Y.; Weng, W.; Peng, J.; Hong, L.; Yang, L.; Toiyama, Y.; Gao, R.; Liu, M.; Yin, M.; Pan, C.; et al. Fusobacterium nucleatum Increases Proliferation of Colorectal Cancer Cells and Tumor Development in Mice by Activating Toll-Like Receptor 4 Signaling to Nuclear Factor-kappaB, and Up-regulating Expression of MicroRNA-21. Gastroenterology 2017, 152, 851–866.
- Gur, C.; Maalouf, N.; Shhadeh, A.; Berhani, O.; Singer, B.B.; Bachrach, G.; Mandelboim, O. Fusobacterium nucleatum supresses anti-tumor immunity by activating CEACAM1. Oncoimmunology 2019, 8, e1581531.
- Brennan, C.A.; Clay, S.L.; Lavoie, S.L.; Bae, S.; Lang, J.K.; Fonseca-Pereira, D.; Rosinski, K.G.; Ou, N.; Glickman, J.N.; Garrett, W.S. Fusobacterium nucleatum drives a pro-inflammatory intestinal microenvironment through metabolite receptor-dependent modulation of IL-17 expression. Gut Microbes 2021, 13, 1987780.
- Loesche, W.J.; Gibbons, R.J. Amino acid fermentation by Fusobacterium nucleatum. Arch. Oral Biol. 1968, 13, 191–202.
- Lavoie, S.; Chun, E.; Bae, S.; Brennan, C.A.; Gallini Comeau, C.A.; Lang, J.K.; Michaud, M.; Hoveyda, H.R.; Fraser, G.L.; Fuller, M.H.; et al. Expression of Free Fatty Acid Receptor 2 by Dendritic Cells Prevents Their Expression of Interleukin 27 and Is Required for Maintenance of Mucosal Barrier and Immune Response Against Colorectal Tumors in Mice. Gastroenterology 2020, 158, 1359–1372.
- Donohoe, D.R.; Holley, D.; Collins, L.B.; Montgomery, S.A.; Whitmore, A.C.; Hillhouse, A.; Curry, K.P.; Renner, S.W.; Greenwalt, A.; Ryan, E.P.; et al. A gnotobiotic mouse model demonstrates that dietary fiber protects against colorectal tumorigenesis in a microbiota- and butyrate-dependent manner. Cancer Discov. 2014, 4, 1387–1397.
- Dong, W.; Liu, L.; Dou, Y.; Xu, M.; Liu, T.; Wang, S.; Zhang, Y.; Deng, B.; Wang, B.; Cao, H. Deoxycholic acid activates epidermal growth factor receptor and promotes intestinal carcinogenesis by ADAM17-dependent ligand release. J. Cell. Mol. Med. 2018, 22, 4263–4273.
- Pai, R.; Tarnawski, A.S.; Tran, T. Deoxycholic acid activates beta-catenin signaling pathway and increases colon cell cancer growth and invasiveness. Mol. Biol. Cell 2004, 15, 2156–2163.
- Fu, T.; Coulter, S.; Yoshihara, E.; Oh, T.G.; Fang, S.; Cayabyab, F.; Zhu, Q.; Zhang, T.; Leblanc, M.; Liu, S.; et al. FXR Regulates Intestinal Cancer Stem Cell Proliferation. Cell 2019, 176, 1098–1112.
- Liu, L.; Yang, M.; Dong, W.; Liu, T.; Song, X.; Gu, Y.; Wang, S.; Liu, Y.; Abla, Z.; Qiao, X.; et al. Gut Dysbiosis and Abnormal Bile Acid Metabolism in Colitis-Associated Cancer. Gastroenterol. Res. Pract. 2021, 2021, 6645970.
- Ternes, D.; Tsenkova, M.; Pozdeev, V.I.; Meyers, M.; Koncina, E.; Atatri, S.; Schmitz, M.; Karta, J.; Schmoetten, M.; Heinken, A.; et al. The gut microbial metabolite formate exacerbates colorectal cancer progression. Nat. Metab. 2022, 4, 458–475.
- Fan, L.; Xu, C.; Ge, Q.; Lin, Y.; Wong, C.C.; Qi, Y.; Ye, B.; Lian, Q.; Zhuo, W.; Si, J.; et al. A. muciniphila Suppresses Colorectal Tumorigenesis by Inducing TLR2/NLRP3-Mediated M1-Like TAMs. Cancer Immunol. Res. 2021, 9, 1111–1124.
- Zegarra Ruiz, D.F.; Kim, D.V.; Norwood, K.; Saldana-Morales, F.B.; Kim, M.; Ng, C.; Callaghan, R.; Uddin, M.; Chang, L.C.; Longman, R.S.; et al. Microbiota manipulation to increase macrophage IL-10 improves colitis and limits colitis-associated colorectal cancer. Gut Microbes 2022, 14, 2119054.
- Yu, A.I.; Zhao, L.; Eaton, K.A.; Ho, S.; Chen, J.; Poe, S.; Becker, J.; Gonzalez, A.; McKinstry, D.; Hasso, M.; et al. Gut Microbiota Modulate CD8 T Cell Responses to Influence Colitis-Associated Tumorigenesis. Cell Rep. 2020, 31, 107471.
- Peuker, K.; Strigli, A.; Tauriello, D.V.F.; Hendricks, A.; von Schonfels, W.; Burmeister, G.; Brosch, M.; Herrmann, A.; Kruger, S.; Nitsche, J.; et al. Microbiota-dependent activation of the myeloid calcineurin-NFAT pathway inhibits B7H3- and B7H4-dependent anti-tumor immunity in colorectal cancer. Immunity 2022, 55, 701–717.
- Overacre-Delgoffe, A.E.; Bumgarner, H.J.; Cillo, A.R.; Burr, A.H.P.; Tometich, J.T.; Bhattacharjee, A.; Bruno, T.C.; Vignali, D.A.A.; Hand, T.W. Microbiota-specific T follicular helper cells drive tertiary lymphoid structures and anti-tumor immunity against colorectal cancer. Immunity 2021, 54, 2812–2824.e2814.
- Noble, A.; Pring, E.T.; Durant, L.; Man, R.; Dilke, S.M.; Hoyles, L.; James, S.A.; Carding, S.R.; Jenkins, J.T.; Knight, S.C. Altered immunity to microbiota, B cell activation and depleted gammadelta/resident memory T cells in colorectal cancer. Cancer Immunol. Immunother. 2022, 71, 2619–2629.
- Yang, J.D.; Hainaut, P.; Gores, G.J.; Amadou, A.; Plymoth, A.; Roberts, L.R. A global view of hepatocellular carcinoma: Trends, risk, prevention and management. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 589–604.
- Huang, D.Q.; El-Serag, H.B.; Loomba, R. Global epidemiology of NAFLD-related HCC: Trends, predictions, risk factors and prevention. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 223–238.
- Park, E.J.; Lee, J.H.; Yu, G.Y.; He, G.; Ali, S.R.; Holzer, R.G.; Osterreicher, C.H.; Takahashi, H.; Karin, M. Dietary and genetic obesity promote liver inflammation and tumorigenesis by enhancing IL-6 and TNF expression. Cell 2010, 140, 197–208.
- Said, I.; Ahad, H.; Said, A. Gut microbiome in non-alcoholic fatty liver disease associated hepatocellular carcinoma: Current knowledge and potential for therapeutics. World J. Gastrointest. Oncol. 2022, 14, 947–958.
- Behary, J.; Raposo, A.E.; Amorim, N.M.L.; Zheng, H.; Gong, L.; McGovern, E.; Chen, J.; Liu, K.; Beretov, J.; Theocharous, C.; et al. Defining the temporal evolution of gut dysbiosis and inflammatory responses leading to hepatocellular carcinoma in Mdr2 −/− mouse model. BMC Microbiol. 2021, 21, 113.
- Ponziani, F.R.; Bhoori, S.; Castelli, C.; Putignani, L.; Rivoltini, L.; Del Chierico, F.; Sanguinetti, M.; Morelli, D.; Paroni Sterbini, F.; Petito, V.; et al. Hepatocellular Carcinoma Is Associated with Gut Microbiota Profile and Inflammation in Nonalcoholic Fatty Liver Disease. Hepatology 2019, 69, 107–120.
- Zheng, R.; Wang, G.; Pang, Z.; Ran, N.; Gu, Y.; Guan, X.; Yuan, Y.; Zuo, X.; Pan, H.; Zheng, J.; et al. Liver cirrhosis contributes to the disorder of gut microbiota in patients with hepatocellular carcinoma. Cancer Med. 2020, 9, 4232–4250.
- Liu, Q.; Li, F.; Zhuang, Y.; Xu, J.; Wang, J.; Mao, X.; Zhang, Y.; Liu, X. Alteration in gut microbiota associated with hepatitis B and non-hepatitis virus related hepatocellular carcinoma. Gut Pathog. 2019, 11, 1.
- Ren, Z.; Li, A.; Jiang, J.; Zhou, L.; Yu, Z.; Lu, H.; Xie, H.; Chen, X.; Shao, L.; Zhang, R.; et al. Gut microbiome analysis as a tool towards targeted non-invasive biomarkers for early hepatocellular carcinoma. Gut 2019, 68, 1014–1023.
- Grat, M.; Wronka, K.M.; Krasnodebski, M.; Masior, L.; Lewandowski, Z.; Kosinska, I.; Grat, K.; Stypulkowski, J.; Rejowski, S.; Wasilewicz, M.; et al. Profile of Gut Microbiota Associated with the Presence of Hepatocellular Cancer in Patients with Liver Cirrhosis. Transplant. Proc. 2016, 48, 1687–1691.
- Ohtani, N.; Hara, E. Gut-liver axis-mediated mechanism of liver cancer: A special focus on the role of gut microbiota. Cancer Sci. 2021, 112, 4433–4443.
- Loo, T.M.; Kamachi, F.; Watanabe, Y.; Yoshimoto, S.; Kanda, H.; Arai, Y.; Nakajima-Takagi, Y.; Iwama, A.; Koga, T.; Sugimoto, Y.; et al. Gut Microbiota Promotes Obesity-Associated Liver Cancer through PGE(2)-Mediated Suppression of Antitumor Immunity. Cancer Discov. 2017, 7, 522–538.
- Golonka, R.M.; Xiao, X.; Abokor, A.A.; Joe, B.; Vijay-Kumar, M. Altered nutrient status reprograms host inflammation and metabolic health via gut microbiota. J. Nutr. Biochem. 2020, 80, 108360.
- Hosseinkhani, F.; Heinken, A.; Thiele, I.; Lindenburg, P.W.; Harms, A.C.; Hankemeier, T. The contribution of gut bacterial metabolites in the human immune signaling pathway of non-communicable diseases. Gut Microbes 2021, 13, 1882927.
- Yue, C.; Yang, X.; Li, J.; Chen, X.; Zhao, X.; Chen, Y.; Wen, Y. Trimethylamine N-oxide prime NLRP3 inflammasome via inhibiting ATG16L1-induced autophagy in colonic epithelial cells. Biochem. Biophys. Res. Commun. 2017, 490, 541–551.
- Wang, Z.; Klipfell, E.; Bennett, B.J.; Koeth, R.; Levison, B.S.; Dugar, B.; Feldstein, A.E.; Britt, E.B.; Fu, X.; Chung, Y.M.; et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 2011, 472, 57–63.
- Guasti, L.; Galliazzo, S.; Molaro, M.; Visconti, E.; Pennella, B.; Gaudio, G.V.; Lupi, A.; Grandi, A.M.; Squizzato, A. TMAO as a biomarker of cardiovascular events: A systematic review and meta-analysis. Intern. Emerg. Med. 2021, 16, 201–207.
- Fang, C.; Zuo, K.; Jiao, K.; Zhu, X.; Fu, Y.; Zhong, J.; Xu, L.; Yang, X. PAGln, an Atrial Fibrillation-Linked Gut Microbial Metabolite, Acts as a Promoter of Atrial Myocyte Injury. Biomolecules 2022, 12, 1120.
- Nemet, I.; Saha, P.P.; Gupta, N.; Zhu, W.; Romano, K.A.; Skye, S.M.; Cajka, T.; Mohan, M.L.; Li, L.; Wu, Y.; et al. A Cardiovascular Disease-Linked Gut Microbial Metabolite Acts via Adrenergic Receptors. Cell 2020, 180, 862–877.
- Bell, K.J.; Saad, S.; Tillett, B.J.; McGuire, H.M.; Bordbar, S.; Yap, Y.A.; Nguyen, L.T.; Wilkins, M.R.; Corley, S.; Brodie, S.; et al. Metabolite-based dietary supplementation in human type 1 diabetes is associated with microbiota and immune modulation. Microbiome 2022, 10, 9.
- Marino, E.; Richards, J.L.; McLeod, K.H.; Stanley, D.; Yap, Y.A.; Knight, J.; McKenzie, C.; Kranich, J.; Oliveira, A.C.; Rossello, F.J.; et al. Gut microbial metabolites limit the frequency of autoimmune T cells and protect against type 1 diabetes. Nat. Immunol. 2017, 18, 552–562.
- Girdhar, K.; Huang, Q.; Chow, I.T.; Vatanen, T.; Brady, C.; Raisingani, A.; Autissier, P.; Atkinson, M.A.; Kwok, W.W.; Kahn, C.R.; et al. A gut microbial peptide and molecular mimicry in the pathogenesis of type 1 diabetes. Proc. Natl. Acad. Sci. USA 2022, 119, e2120028119.
- Lamichhane, S.; Sen, P.; Dickens, A.M.; Alves, M.A.; Harkonen, T.; Honkanen, J.; Vatanen, T.; Xavier, R.J.; Hyotylainen, T.; Knip, M.; et al. Dysregulation of secondary bile acid metabolism precedes islet autoimmunity and type 1 diabetes. Cell Rep. Med. 2022, 3, 100762.
- Pedersen, H.K.; Gudmundsdottir, V.; Nielsen, H.B.; Hyotylainen, T.; Nielsen, T.; Jensen, B.A.; Forslund, K.; Hildebrand, F.; Prifti, E.; Falony, G.; et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature 2016, 535, 376–381.
- Zhou, M.; Shao, J.; Wu, C.Y.; Shu, L.; Dong, W.; Liu, Y.; Chen, M.; Wynn, R.M.; Wang, J.; Wang, J.; et al. Targeting BCAA Catabolism to Treat Obesity-Associated Insulin Resistance. Diabetes 2019, 68, 1730–1746.
- Bodogai, M.; O′Connell, J.; Kim, K.; Kim, Y.; Moritoh, K.; Chen, C.; Gusev, F.; Vaughan, K.; Shulzhenko, N.; Mattison, J.A.; et al. Commensal bacteria contribute to insulin resistance in aging by activating innate B1a cells. Sci. Transl. Med. 2018, 10, eaat4271.
- Huang, J.P.; Cheng, M.L.; Hung, C.Y.; Wang, C.H.; Hsieh, P.S.; Shiao, M.S.; Chen, J.K.; Li, D.E.; Hung, L.M. Docosapentaenoic acid and docosahexaenoic acid are positively associated with insulin sensitivity in rats fed high-fat and high-fructose diets. J. Diabetes 2017, 9, 936–946.
- Wang, H.; Lu, Y.; Yan, Y.; Tian, S.; Zheng, D.; Leng, D.; Wang, C.; Jiao, J.; Wang, Z.; Bai, Y. Promising Treatment for Type 2 Diabetes: Fecal Microbiota Transplantation Reverses Insulin Resistance and Impaired Islets. Front. Cell. Infect. Microbiol. 2019, 9, 455.
- Deng, X.; Zhang, C.; Wang, P.; Wei, W.; Shi, X.; Wang, P.; Yang, J.; Wang, L.; Tang, S.; Fang, Y.; et al. Cardiovascular Benefits of Empagliflozin Are Associated with Gut Microbiota and Plasma Metabolites in Type 2 Diabetes. J. Clin. Endocrinol. Metab. 2022, 107, 1888–1896.
- Montandon, S.A.; Jornayvaz, F.R. Effects of Antidiabetic Drugs on Gut Microbiota Composition. Genes 2017, 8, 250.
- Wu, H.; Esteve, E.; Tremaroli, V.; Khan, M.T.; Caesar, R.; Manneras-Holm, L.; Stahlman, M.; Olsson, L.M.; Serino, M.; Planas-Felix, M.; et al. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat. Med. 2017, 23, 850–858.
- Li, J.; Zhao, F.; Wang, Y.; Chen, J.; Tao, J.; Tian, G.; Wu, S.; Liu, W.; Cui, Q.; Geng, B.; et al. Gut microbiota dysbiosis contributes to the development of hypertension. Microbiome 2017, 5, 14.
- Adnan, S.; Nelson, J.W.; Ajami, N.J.; Venna, V.R.; Petrosino, J.F.; Bryan, R.M., Jr.; Durgan, D.J. Alterations in the gut microbiota can elicit hypertension in rats. Physiol. Genom. 2017, 49, 96–104.
- Marques, F.Z.; Nelson, E.; Chu, P.Y.; Horlock, D.; Fiedler, A.; Ziemann, M.; Tan, J.K.; Kuruppu, S.; Rajapakse, N.W.; El-Osta, A.; et al. High-Fiber Diet and Acetate Supplementation Change the Gut Microbiota and Prevent the Development of Hypertension and Heart Failure in Hypertensive Mice. Circulation 2017, 135, 964–977.
- Santisteban, M.M.; Qi, Y.; Zubcevic, J.; Kim, S.; Yang, T.; Shenoy, V.; Cole-Jeffrey, C.T.; Lobaton, G.O.; Stewart, D.C.; Rubiano, A.; et al. Hypertension-Linked Pathophysiological Alterations in the Gut. Circ. Res. 2017, 120, 312–323.
- Karbach, S.H.; Schonfelder, T.; Brandao, I.; Wilms, E.; Hormann, N.; Jackel, S.; Schuler, R.; Finger, S.; Knorr, M.; Lagrange, J.; et al. Gut Microbiota Promote Angiotensin II-Induced Arterial Hypertension and Vascular Dysfunction. J. Am. Heart Assoc. 2016, 5, e003698.
- Avery, E.G.; Bartolomaeus, H.; Rauch, A.; Chen, C.Y.; N′Diaye, G.; Lober, U.; Bartolomaeus, T.U.P.; Fritsche-Guenther, R.; Rodrigues, A.F.; Yarritu, A.; et al. Quantifying the impact of gut microbiota on inflammation and hypertensive organ damage. Cardiovasc. Res. 2022.
- Joe, B.; McCarthy, C.G.; Edwards, J.M.; Cheng, X.; Chakraborty, S.; Yang, T.; Golonka, R.M.; Mell, B.; Yeo, J.Y.; Bearss, N.R.; et al. Microbiota Introduced to Germ-Free Rats Restores Vascular Contractility and Blood Pressure. Hypertension 2020, 76, 1847–1855.
- Abboud, F.M.; Cicha, M.Z.; Ericsson, A.; Chapleau, M.W.; Singh, M.V. Altering Early Life Gut Microbiota Has Long-Term Effect on Immune System and Hypertension in Spontaneously Hypertensive Rats. Front. Physiol. 2021, 12, 752924.
- Jiang, S.; Shui, Y.; Cui, Y.; Tang, C.; Wang, X.; Qiu, X.; Hu, W.; Fei, L.; Li, Y.; Zhang, S.; et al. Gut microbiota dependent trimethylamine N-oxide aggravates angiotensin II-induced hypertension. Redox Biol. 2021, 46, 102115.
- Ferguson, J.F.; Aden, L.A.; Barbaro, N.R.; Van Beusecum, J.P.; Xiao, L.; Simmons, A.J.; Warden, C.; Pasic, L.; Himmel, L.E.; Washington, M.K.; et al. High dietary salt-induced dendritic cell activation underlies microbial dysbiosis-associated hypertension. JCI Insight 2019, 5, e126241.
- Chakraborty, S.; Galla, S.; Cheng, X.; Yeo, J.Y.; Mell, B.; Singh, V.; Yeoh, B.; Saha, P.; Mathew, A.V.; Vijay-Kumar, M.; et al. Salt-Responsive Metabolite, beta-Hydroxybutyrate, Attenuates Hypertension. Cell Rep. 2018, 25, 677–689.
- Wilck, N.; Matus, M.G.; Kearney, S.M.; Olesen, S.W.; Forslund, K.; Bartolomaeus, H.; Haase, S.; Mahler, A.; Balogh, A.; Marko, L.; et al. Salt-responsive gut commensal modulates T(H)17 axis and disease. Nature 2017, 551, 585–589.
- Lee, Y.H. Causal association of gut microbiome on the risk of rheumatoid arthritis: A Mendelian randomisation study. Ann. Rheum. Dis. 2022, 81, e3.
- Kishikawa, T.; Maeda, Y.; Nii, T.; Motooka, D.; Matsumoto, Y.; Matsushita, M.; Matsuoka, H.; Yoshimura, M.; Kawada, S.; Teshigawara, S.; et al. Metagenome-wide association study of gut microbiome revealed novel aetiology of rheumatoid arthritis in the Japanese population. Ann. Rheum. Dis. 2020, 79, 103–111.
- Pianta, A.; Arvikar, S.; Strle, K.; Drouin, E.E.; Wang, Q.; Costello, C.E.; Steere, A.C. Evidence of the Immune Relevance of Prevotella copri, a Gut Microbe, in Patients with Rheumatoid Arthritis. Arthritis Rheumatol. 2017, 69, 964–975.
- Li, Y.; Zhang, S.X.; Yin, X.F.; Zhang, M.X.; Qiao, J.; Xin, X.H.; Chang, M.J.; Gao, C.; Li, Y.F.; Li, X.F. The Gut Microbiota and Its Relevance to Peripheral Lymphocyte Subpopulations and Cytokines in Patients with Rheumatoid Arthritis. J. Immunol. Res. 2021, 2021, 6665563.
- Teng, F.; Klinger, C.N.; Felix, K.M.; Bradley, C.P.; Wu, E.; Tran, N.L.; Umesaki, Y.; Wu, H.J. Gut Microbiota Drive Autoimmune Arthritis by Promoting Differentiation and Migration of Peyer′s Patch T Follicular Helper Cells. Immunity 2016, 44, 875–888.
- Bates, N.A.; Li, A.; Fan, T.; Cutcliffe, M.P.; Dagenet, C.B.; Sleiman, K.C.; Ma, H.; Tahsin, S.; Garrett, C.S.; Altemus, J.; et al. Gut Commensal Segmented Filamentous Bacteria Fine-Tune T Follicular Regulatory Cells to Modify the Severity of Systemic Autoimmune Arthritis. J. Immunol. 2021, 206, 941–952.
- Teng, F.; Felix, K.M.; Bradley, C.P.; Naskar, D.; Ma, H.; Raslan, W.A.; Wu, H.J. The impact of age and gut microbiota on Th17 and Tfh cells in K/BxN autoimmune arthritis. Arthritis Res. Ther. 2017, 19, 188.
- Block, K.E.; Zheng, Z.; Dent, A.L.; Kee, B.L.; Huang, H. Gut Microbiota Regulates K/BxN Autoimmune Arthritis through Follicular Helper T but Not Th17 Cells. J. Immunol. 2016, 196, 1550–1557.
- Simpkins, D.A.; Downton, P.; Gray, K.J.; Dickson, S.H.; Maidstone, R.J.; Konkel, J.E.; Hepworth, M.R.; Ray, D.W.; Bechtold, D.A.; Gibbs, J.E. Consequences of collagen induced inflammatory arthritis on circadian regulation of the gut microbiome. FASEB J. 2023, 37, e22704.
- He, J.; Chu, Y.; Li, J.; Meng, Q.; Liu, Y.; Jin, J.; Wang, Y.; Wang, J.; Huang, B.; Shi, L.; et al. Intestinal butyrate-metabolizing species contribute to autoantibody production and bone erosion in rheumatoid arthritis. Sci. Adv. 2022, 8, eabm1511.
- Takahashi, D.; Hoshina, N.; Kabumoto, Y.; Maeda, Y.; Suzuki, A.; Tanabe, H.; Isobe, J.; Yamada, T.; Muroi, K.; Yanagisawa, Y.; et al. Microbiota-derived butyrate limits the autoimmune response by promoting the differentiation of follicular regulatory T cells. EBioMedicine 2020, 58, 102913.
- Rosser, E.C.; Piper, C.J.M.; Matei, D.E.; Blair, P.A.; Rendeiro, A.F.; Orford, M.; Alber, D.G.; Krausgruber, T.; Catalan, D.; Klein, N.; et al. Microbiota-Derived Metabolites Suppress Arthritis by Amplifying Aryl-Hydrocarbon Receptor Activation in Regulatory B Cells. Cell Metab. 2020, 31, 837–851.
- Sun, H.; Guo, Y.; Wang, H.; Yin, A.; Hu, J.; Yuan, T.; Zhou, S.; Xu, W.; Wei, P.; Yin, S.; et al. Gut commensal Parabacteroides distasonis alleviates inflammatory arthritis. Gut 2023.
- Adeyeye, T.E.; Yeung, E.H.; McLain, A.C.; Lin, S.; Lawrence, D.A.; Bell, E.M. Wheeze and Food Allergies in Children Born via Cesarean Delivery: The Upstate KIDS Study. Am. J. Epidemiol. 2019, 188, 355–362.
- Loh, W.; Tang, M.L.K. The Epidemiology of Food Allergy in the Global Context. Int J. Environ. Res. Public Health 2018, 15, 2043.
- Leung, A.S.Y.; Wong, G.W.K.; Tang, M.L.K. Food allergy in the developing world. J. Allergy Clin. Immunol. 2018, 141, 76–78.
- Borbet, T.C.; Pawline, M.B.; Zhang, X.; Wipperman, M.F.; Reuter, S.; Maher, T.; Li, J.; Iizumi, T.; Gao, Z.; Daniele, M.; et al. Influence of the early-life gut microbiota on the immune responses to an inhaled allergen. Mucosal Immunol. 2022, 15, 1000–1011.
- Zhang, Q.; Cheng, L.; Wang, J.; Hao, M.; Che, H. Antibiotic-Induced Gut Microbiota Dysbiosis Damages the Intestinal Barrier, Increasing Food Allergy in Adult Mice. Nutrients 2021, 13, 3315.
- Hussain, M.; Bonilla-Rosso, G.; Kwong Chung, C.K.C.; Bariswyl, L.; Rodriguez, M.P.; Kim, B.S.; Engel, P.; Noti, M. High dietary fat intake induces a microbiota signature that promotes food allergy. J. Allergy Clin. Immunol. 2019, 144, 157–170.
- Zimmermann, P.; Messina, N.; Mohn, W.W.; Finlay, B.B.; Curtis, N. Association between the intestinal microbiota and allergic sensitization, eczema, and asthma: A systematic review. J. Allergy Clin. Immunol. 2019, 143, 467–485.
- McCauley, K.E.; Rackaityte, E.; LaMere, B.; Fadrosh, D.W.; Fujimura, K.E.; Panzer, A.R.; Lin, D.L.; Lynch, K.V.; Halkias, J.; Mendoza, V.F.; et al. Heritable vaginal bacteria influence immune tolerance and relate to early-life markers of allergic sensitization in infancy. Cell Rep. Med. 2022, 3, 100713.
- Okba, A.M.; Saber, S.M.; Abdel-Rehim, A.S.; Amin, M.M.; Mohamed, D.A. Fecal microbiota profile in atopic asthmatic adult patients. Eur. Ann. Allergy Clin. Immunol. 2018, 50, 117–124.
- Lundstrom, S.L.; Yang, J.; Kallberg, H.J.; Thunberg, S.; Gafvelin, G.; Haeggstrom, J.Z.; Gronneberg, R.; Grunewald, J.; van Hage, M.; Hammock, B.D.; et al. Allergic asthmatics show divergent lipid mediator profiles from healthy controls both at baseline and following birch pollen provocation. PLoS ONE 2012, 7, e33780.
- Fujimura, K.E.; Sitarik, A.R.; Havstad, S.; Lin, D.L.; Levan, S.; Fadrosh, D.; Panzer, A.R.; LaMere, B.; Rackaityte, E.; Lukacs, N.W.; et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat. Med. 2016, 22, 1187–1191.
- Levan, S.R.; Stamnes, K.A.; Lin, D.L.; Panzer, A.R.; Fukui, E.; McCauley, K.; Fujimura, K.E.; McKean, M.; Ownby, D.R.; Zoratti, E.M.; et al. Elevated faecal 12,13-diHOME concentration in neonates at high risk for asthma is produced by gut bacteria and impedes immune tolerance. Nat. Microbiol. 2019, 4, 1851–1861.
- Abdulla, O.A.; Neamah, W.; Sultan, M.; Alghetaa, H.K.; Singh, N.; Busbee, P.B.; Nagarkatti, M.; Nagarkatti, P. The Ability of AhR Ligands to Attenuate Delayed Type Hypersensitivity Reaction Is Associated with Alterations in the Gut Microbiota. Front. Immunol. 2021, 12, 684727.
- Roduit, C.; Frei, R.; Ferstl, R.; Loeliger, S.; Westermann, P.; Rhyner, C.; Schiavi, E.; Barcik, W.; Rodriguez-Perez, N.; Wawrzyniak, M.; et al. High levels of butyrate and propionate in early life are associated with protection against atopy. Allergy 2019, 74, 799–809.
- Cait, A.; Hughes, M.R.; Antignano, F.; Cait, J.; Dimitriu, P.A.; Maas, K.R.; Reynolds, L.A.; Hacker, L.; Mohr, J.; Finlay, B.B.; et al. Microbiome-driven allergic lung inflammation is ameliorated by short-chain fatty acids. Mucosal Immunol. 2018, 11, 785–795.
- Thorburn, A.N.; McKenzie, C.I.; Shen, S.; Stanley, D.; Macia, L.; Mason, L.J.; Roberts, L.K.; Wong, C.H.; Shim, R.; Robert, R.; et al. Evidence that asthma is a developmental origin disease influenced by maternal diet and bacterial metabolites. Nat. Commun. 2015, 6, 7320.
- Tan, J.; McKenzie, C.; Vuillermin, P.J.; Goverse, G.; Vinuesa, C.G.; Mebius, R.E.; Macia, L.; Mackay, C.R. Dietary Fiber and Bacterial SCFA Enhance Oral Tolerance and Protect against Food Allergy through Diverse Cellular Pathways. Cell Rep. 2016, 15, 2809–2824.
- Arifuzzaman, M.; Won, T.H.; Li, T.T.; Yano, H.; Digumarthi, S.; Heras, A.F.; Zhang, W.; Parkhurst, C.N.; Kashyap, S.; Jin, W.B.; et al. Inulin fibre promotes microbiota-derived bile acids and type 2 inflammation. Nature 2022, 611, 578–584.
- Seki, D.; Mayer, M.; Hausmann, B.; Pjevac, P.; Giordano, V.; Goeral, K.; Unterasinger, L.; Klebermass-Schrehof, K.; De Paepe, K.; Van de Wiele, T.; et al. Aberrant gut-microbiota-immune-brain axis development in premature neonates with brain damage. Cell Host Microbe 2021, 29, 1558–1572.
- Giuffre, M.; Moretti, R.; Campisciano, G.; da Silveira, A.B.M.; Monda, V.M.; Comar, M.; Di Bella, S.; Antonello, R.M.; Luzzati, R.; Croce, L.S. You Talking to Me? Says the Enteric Nervous System (ENS) to the Microbe. How Intestinal Microbes Interact with the ENS. J. Clin. Med. 2020, 9, 3705.
- Ouabbou, S.; He, Y.; Butler, K.; Tsuang, M. Inflammation in Mental Disorders: Is the Microbiota the Missing Link? Neurosci. Bull. 2020, 36, 1071–1084.
- Slyepchenko, A.; Carvalho, A.F.; Cha, D.S.; Kasper, S.; McIntyre, R.S. Gut emotions—Mechanisms of action of probiotics as novel therapeutic targets for depression and anxiety disorders. CNS Neurol. Disord. Drug Targets 2014, 13, 1770–1786.
- Zhang, X.; Hou, Y.; Li, Y.; Wei, W.; Cai, X.; Shao, H.; Yuan, Y.; Zheng, X. Taxonomic and Metabolic Signatures of Gut Microbiota for Assessing the Severity of Depression and Anxiety in Major Depressive Disorder Patients. Neuroscience 2022, 496, 179–189.
- Galley, J.D.; Mashburn-Warren, L.; Blalock, L.C.; Lauber, C.L.; Carroll, J.E.; Ross, K.M.; Hobel, C.; Coussons-Read, M.; Dunkel Schetter, C.; Gur, T.L. Maternal anxiety, depression and stress affects offspring gut microbiome diversity and bifidobacterial abundances. Brain Behav. Immun. 2023, 107, 253–264.
- O′Neill, S.M.; Curran, E.A.; Dalman, C.; Kenny, L.C.; Kearney, P.M.; Clarke, G.; Cryan, J.F.; Dinan, T.G.; Khashan, A.S. Birth by Caesarean Section and the Risk of Adult Psychosis: A Population-Based Cohort Study. Schizophr. Bull. 2016, 42, 633–641.
- Morais, L.H.; Golubeva, A.V.; Casey, S.; Scott, K.A.; Ramos Costa, A.P.; Moloney, G.M.; Dinan, T.G.; Cryan, J.F. Early-life oxytocin attenuates the social deficits induced by caesarean-section delivery in the mouse. Neuropsychopharmacology 2021, 46, 1958–1968.
- Lynch, C.M.K.; Cowan, C.S.M.; Bastiaanssen, T.F.S.; Moloney, G.M.; Theune, N.; van de Wouw, M.; Florensa Zanuy, E.; Ventura-Silva, A.P.; Codagnone, M.G.; Villalobos-Manriquez, F.; et al. Critical windows of early-life microbiota disruption on behaviour, neuroimmune function, and neurodevelopment. Brain Behav. Immun. 2022, 108, 309–327.
- Li, J.; Pu, F.; Peng, C.; Wang, Y.; Zhang, Y.; Wu, S.; Wang, S.; Shen, X.; Li, Y.; Cheng, R.; et al. Antibiotic cocktail-induced gut microbiota depletion in different stages could cause host cognitive impairment and emotional disorders in adulthood in different manners. Neurobiol. Dis. 2022, 170, 105757.
- Cuskelly, A.; Hoedt, E.C.; Harms, L.; Talley, N.J.; Tadros, M.A.; Keely, S.; Hodgson, D.M. Neonatal immune challenge influences the microbiota and behaviour in a sexually dimorphic manner. Brain Behav. Immun. 2022, 103, 232–242.
- Lee, J.; Venna, V.R.; Durgan, D.J.; Shi, H.; Hudobenko, J.; Putluri, N.; Petrosino, J.; McCullough, L.D.; Bryan, R.M. Young versus aged microbiota transplants to germ-free mice: Increased short-chain fatty acids and improved cognitive performance. Gut Microbes 2020, 12, 1814107.
- Leonardi, I.; Gao, I.H.; Lin, W.Y.; Allen, M.; Li, X.V.; Fiers, W.D.; De Celie, M.B.; Putzel, G.G.; Yantiss, R.K.; Johncilla, M.; et al. Mucosal fungi promote gut barrier function and social behavior via Type 17 immunity. Cell 2022, 185, 831–846.
