In polymer science, the backbone chain of a polymer is the longest series of covalently bonded atoms that together create the continuous chain of the molecule. This science is subdivided into the study of organic polymers, which consist of a carbon backbone, and inorganic polymers which have backbones containing only main group elements. In biochemistry, organic backbone chains make up the primary structure of macromolecules. The backbones of these biological macromolecules consist of central chains of covalently bonded atoms. The characteristics and order of the monomer residues in the backbone make a map for the complex structure of biological polymers (see Biomolecular structure). The backbone is, therefore, directly related to biological molecules’ function. The macromolecules within the body can be divided into four main subcategories, each of which are involved in very different and important biological processes: proteins, carbohydrates, lipids, and nucleic acids. Each of these molecules has a different backbone and consists of different monomers each with distinctive residues and functionalities. This is the driving factor of their different structures and functions in the body. Although lipids have a "backbone," they are not true biological polymers as their backbone is a three carbon molecule, glycerol, with longer substituent "side chains." For this reason, only proteins, carbohydrates, and nucleic acids should be considered as biological macromolecules with polymeric backbones.
- glycerol
- macromolecules
- biomolecular
1. Characteristics
1.1. Polymer Chemistry
The character of the backbone chain depends on the type of polymerization: in step-growth polymerization, the monomer moiety becomes the backbone, and thus the backbone is typically functional. These include polythiophenes or low band gap polymers in organic semiconductors.[1] In chain-growth polymerization, typically applied for alkenes, the backbone is not functional, but bears the functional side chains or pendant groups.
The character of the backbone, i.e. its flexibility, determines the thermal properties of the polymer (such as the glass transition temperature). For example, in polysiloxanes (silicone), the backbone chain is very flexible, which results in a very low glass transition temperature of −123 °C (−189 °F; 150 K).[2] The polymers with rigid backbones are prone to crystallization (e.g. polythiophenes) in thin films and in solution. Crystallization in its turn affects the optical properties of the polymers, its optical band gap and electronic levels.[3]
1.2. Biochemistry
There are some similarities and many differences inherent in the character of biopolymer backbones. The backbone of each of the three biological polymers; proteins, carbohydrates, and nucleic acids, is formed through a net condensation reaction. In a condensation reaction, monomers are covalently connected along with the loss of some small molecule, most commonly water.[4] Because they are polymerized through complex enzymatic mechanisms, none of the biopolymers' backbones are formed through the elimination of water but through the elimination of other small biological molecules. Each of these biopolymers can be characterized as either a heteropolymer, meaning it consists of more than one monomer ordered in the backbone chain, or a homopolymer, which consists of just one repeating monomer. Polypeptides and nucleic acids are very commonly heteropolymers whereas common carbohydrate macromolecules such as glycogen can be homopolymers. This is because the chemical differences of peptide and nucleotide monomers determines the biological function of their polymers whereas common carbohydrate monomers have one general function such as for energy storage and delivery.
2. Overview of Common Backbones
2.1. Polymer Chemistry
- saturated alkane (typical for vinyl polymers)
- step-growth polymers (polyaniline, polythiophene, PEDOT) backbone. These often have derivatized heterocycles as monomers, such as thiophenes, diazoles or pyrroles.
- fullerene backbone[5]
2.2. Biology
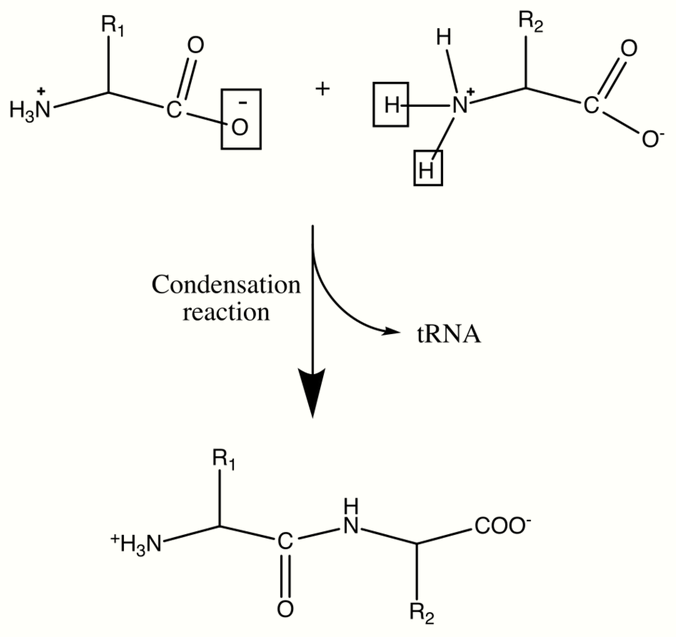
Proteins (Polypeptides)
Proteins are important biological molecules and play an integral role in the structure and function of viruses, bacteria, and eukaryotic cells. Their backbones are characterized by amide linkages formed by the polymerization between amino and carboxylic acid groups attached to the alpha carbon of each of the twenty amino acids. These amino acid sequences are translated from cellular mRNAs by ribosomes in the cytoplasm of the cell.[6] The ribosomes have enzymatic activity which directs the condensation reaction forming the amide linkage between each successive amino acid. This happens during a biological process known as translation. In this enzymatic mechanism a covalently bonded tRNA shuttle acts as the leaving group for the condensation reaction. The newly liberated tRNA can "pick up" another peptide and continuously participate in this reaction.[7] The sequence of the amino acids in the polypeptide backbone is known as the primary structure of the protein. This primary structure leads to folding of the protein into the secondary structure, formed by hydrogen bonding between the carbonyl oxygens and amine hydrogens in the backbone. Further interactions between residues of the individual amino acids form the protein's tertiary structure. For this reason, the primary structure of the amino acids in the polypeptide backbone is the map of the final structure of a protein, and it therefore indicates its biological function.[8][9] Spatial positions of backbone atoms can be reconstructed from the positions of alpha carbons using computational tools for the backbone reconstruction.[10]
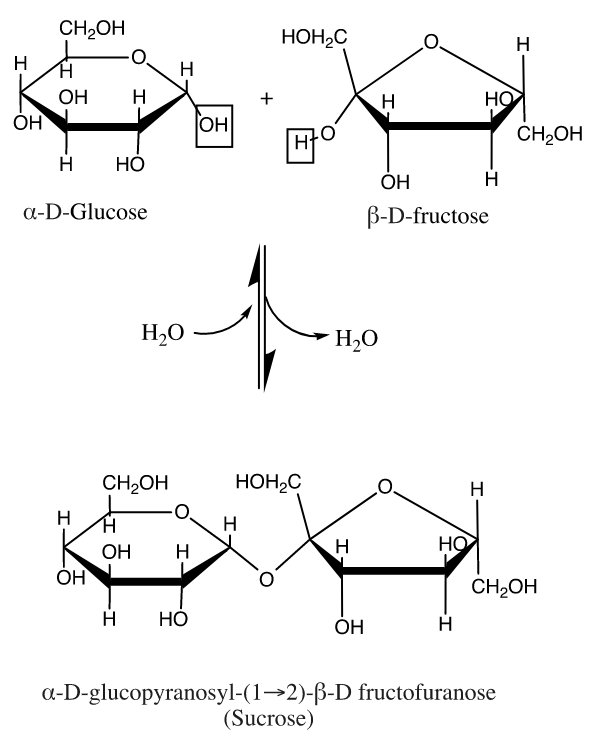
Carbohydrates
Carbohydrates have many roles in the body including functioning as structural units, enzyme cofactors and cell surface recognition sites. Their most prevalent role is in energy storage and delivery in cellular metabolic pathways. The most simple carbohydrates are single sugar residues called monosaccharides like glucose, our body's energy delivery molecule. Oligosaccharides (up to 10 residues) and polysaccharides (up to about 50,000 residues) consist of saccharide residues bonded in a backbone chain, which is characterized by an ether bond known as a glycosidic linkage. In the body's formation of glycogen, the energy storage polymer, this glycosidic linkage is formed by the enzyme glycogen synthase. The mechanism of this enzymatically driven condensation reaction is not well studied but it is known that the molecule UDP acts as an intermediary linker and is lost in the synthesis.[11] These backbone chains can be unbranched (containing one linear chain) or branched (containing multiple chains). The glycosidic linkages are designated as alpha or beta depending on the relative stereochemistry of the anomeric (or most oxidized) carbon. In a Fischer Projection, if the glycosidic linkage is on the same side or face as carbon 6 of a common biological saccharide, the carbohydrate is designated as beta and if the linkage is on the opposite side it is designated as alpha. In a traditional "chair structure" projection, if the linkage is on the same plane (equatorial or axial) as carbon 6 it is designated as beta and on the opposite plane it is designated as alpha. This is exemplified in sucrose (table sugar) which contains a linkage that is alpha to glucose and beta to fructose. Generally, carbohydrates which our bodies break down are alpha-linked (example: glycogen) and those which have structural function are beta-linked (example: cellulose).[9][12]
Nucleic Acids
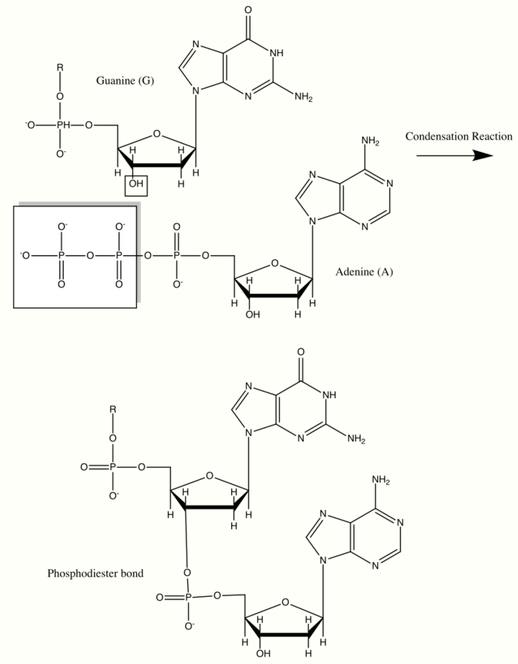
Deoxyribonucleic acid (DNA) and ribonucleic acid (RNA) are of great importance because they code for the production of all cellular proteins. They are made up of monomers called nucleotides which consist of an organic base: A, G, C and T or U, a pentose sugar, and a phosphate group. They have backbones in which the 3’ carbon of the ribose sugar is connected to the phosphate group via a phosphodiester bond. This bond is formed through with the help of a class of cellular enzymes called polymerases. In this enzymatically driven condensation reaction, all incoming nucleotides have a triphosphorylated ribose which loses a pyrophosphate group to form the inherent phosphodiester bond. This reaction is driven by the large negative free energy change associated with the release of pyrophosphate. The sequence of bases in the nucleic acid backbone is also known as the primary structure. Nucleic acids can be millions of nucleotides long thus leading to the genetic diversity of life. The bases stick out from the pentose-phosphate polymer backbone in DNA and are hydrogen bonded in pairs to their complementary partners (A with T and G with C). This creates a double helix with pentose phosphate backbones on either side, thus forming a secondary structure.[9][13][14]
The content is sourced from: https://handwiki.org/wiki/Chemistry:Backbone_chain
References
- Budgaard, Eva; Krebs, Frederik (2006). "Low band gap polymers for organic photovoltaics". Solar Energy Materials and Solar Cells 91 (11): 954–985. doi:10.1016/j.solmat.2007.01.015. https://dx.doi.org/10.1016%2Fj.solmat.2007.01.015
- "Polymers". http://courses.chem.psu.edu/chem112/materials/polymers.html.
- Brabec, C.J.; Winder, C.; Scharber, M.C; Sarıçiftçi, S.N.; Hummelen, J.C.; Svensson, M.; Andersson, M.R. (2001). "Influence of disorder on the photoinduced excitations in phenyl substituted polythiophenes". Journal of Chemical Physics 115 (15): 7235. doi:10.1063/1.1404984. Bibcode: 2001JChPh.115.7235B. https://pure.rug.nl/ws/files/6636890/2001BrabecJChemPhys.pdf.
- IUPAC, Compendium of Chemical Terminology, 2nd ed. (the "Gold Book") (1997). Online corrected version: (2006–) "condensation reaction". doi:10.1351/goldbook.C01238 http://goldbook.iupac.org/C01238.html
- Hirsch, Andreas (1993). "Fullerene polymers". Advanced Materials 5 (11): 859–861. doi:10.1002/adma.19930051116. https://dx.doi.org/10.1002%2Fadma.19930051116
- Noller, HF (2017). "The parable of the caveman and the Ferrari: protein synthesis and the RNA world". Phil. Trans. R. Soc. B 372 (1716): 20160187. doi:10.1098/rstb.2016.0187. PMID 28138073. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=5311931
- Weinger, Joshua (2006). "Participation of the tRNA A76 hydroxyl groups throughout translation". Biochemistry 45 (19): 5939–5948. doi:10.1021/bi060183n. PMID 16681365. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=2522371
- "3.2 Primary Structure: Amino Acids Are Linked by Peptide Bonds to Form Polypeptide Chains". Biochemistry (5th ed.). W.H. Freeman. 2002. NBK22364. ISBN 0-7167-3051-0. https://www.ncbi.nlm.nih.gov/books/NBK22364/.
- Voet, Donald; Voet, Judith G.; Pratt, Charlotte W. (2016). Fundamentals of Biochemistry: Life at the Molecular Level (5th ed.). Wiley. ISBN 978-1-118-91840-1. https://books.google.com/books?id=9T7hCgAAQBAJ. V
- Badaczewska-Dawid, Aleksandra E.; Kolinski, Andrzej; Kmiecik, Sebastian (2020). "Computational reconstruction of atomistic protein structures from coarse-grained models". Computational and Structural Biotechnology Journal 18: 162–176. doi:10.1016/j.csbj.2019.12.007. ISSN 2001-0370. PMID 31969975. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=6961067
- Buschiazzo, Alejandro (2004). "Crystal structure of glycogen synthase: homologous enzymes catalyze glycogen synthesis and degradation". The EMBO Journal 23 (16): 3196–3205. doi:10.1038/sj.emboj.7600324. PMID 15272305. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=514502
- "Structural Basis of Glycan Diversity". Essentials of Glycobiology (2nd ed.). Cold Spring Harbor Laboratory Press. 2009. ISBN 9780879697709. https://www.ncbi.nlm.nih.gov/books/NBK1955/.
- "DNA Replication Mechanisms". Molecular Biology of the Cell (4th ed.). Garland Science. 2002. NBK26850. ISBN 0-8153-3218-1. https://www.ncbi.nlm.nih.gov/books/NBK26850/.
- "4.1, Structure of Nucleic Acids". Molecular Cell Biology (4th ed.). W.H. Freeman. 2000. NBK21514. ISBN 0-7167-3136-3. https://www.ncbi.nlm.nih.gov/books/NBK21514/.
