β-glucosidases (EC. 3.2.1.21) are enzymes that hydrolyze glucosidic bonds of oligosaccharides, in special disaccharides, such as cellobiose, realizing glucose at the end of the process. They are highly used in second-generation biofuel production.
- enzymes
- bioinformatics
- β-glucosidase
- biofuel
- second-generation biofuel
1. Introduction
In second-generation biofuel production, they act in synergy with other classes of enzymes, such as endo-glucanases and exo-glucanases [2][3], to obtain fermentable sugars from biomass. However, the literature has described that most β-glucosidase enzymes are inhibited by glucose, which has been considered one of the most impacting bottlenecks for high-efficient industrial production [4].
Recently, a more efficient class of β-glucosidases have been described in the literature. They are called glucose-tolerant due to their high resistance to inhibition even in high glucose concentrations [5]. This class of enzymes has excellent potential for use in industrial applications such as biofuel production. Hence, glucose-tolerant β-glucosidase enzymes have been an essential target of several studies to detect mutations that transform non-tolerant enzymes into glucose-tolerant enzymes [6].
Most glucose-tolerant β-glucosidases are obtained from glycoside-hydrolase family 1 (GH1). Below, you can see the sequence of the beta-glucosidase from a metagenome sample collected in the South China Sea. This sequence was obtained in the BETAGDB database (available at http://bioinfo.dcc.ufmg.br/betagdb/). GH1 beta-glucosidases present aproximadelly 400 residues (each letter represents an amino acid residue).
>Yang_Y et al. (2015) tr|D5KX75|D5KX75_9BACT Beta-glucosidase OS=uncultured bacterium PE=3 SV=1 MTKISLPTCSPLLTKEFIYGVATASFQIEGGSAHRLPCIWDTFCDTPGKIADNSNGHVAC DHYNNWKQDIDLIESLGVDAYRLSISWPRVITKSGELNPEGVKFYTDILDELKKRNIKAF VTLYHWDLPQHLEDEGGWLNRETAYAFAHYVDLITLAFGDRVHSYATLNEPFCSAFLGYE IGIHAPGKVGKQYGRKAAHHLLLAHGLAMTVLKQNSPTTLNGIVLNFTPCYSISEDADDI AATAFADDYLNQWYMKPIMDGTYPAIIEQLPSAHLPDIHDGDMAIISQSIDYLGINFYTR QFYKAHPTEIYEPIEPTGPLTDMGWEIYPKSFTELLVTLNNTYTLPPIFITENGAAMPDS YNNGEINDVDRLDYYNSHLNAVHNATEQGVRIDGYFAWSLMDNFEWAEGYLKRFGIVYVD YSTQQRTIKNSGLAYKALISNR |
Another example is the β-glucosidase structures obtained from fungi, such as Humicola grisea and Humicola insolens. The sequence below illustrates the primary structure of a fungi β-glucosidase structure.
>de Giuseppe et al. (2014); Benoliel et al. (2010) tr|O93784|O93784_HUMGT Beta-glucosidase OS=Humicola grisea var. thermoidea GN=bgl4 PE=1 SV=1 PDB=4mdo MSLPPDFKWGFATAAYQIEGSVNEDGRGPSIWDTFCAIPGKIADGSSGAVACDSYKRTKE DIALLKELGANSYRFSISWSRIIPLGGRNDPINQKGIDHYVKFVDDLIEAGITPFITLFH WDLPDALDKRYGGFLNKEEFAADFENYARIMFKAIPKCKHWITFNEPWCSAILGYNTGYF APGHTSDRSKSPVGDSAREPWIVGHNILIAHARAVKAYREDFKPTQGGEIGITLNGDATL PWDPEDPADIEACDRKIEFAISWFADPIYFGKYPDSMRKQLGDRLPEFTPEEVALVKGSN DFYGMNHYTANYIKHKTGVPPEDDFLGNLETLFYNKYGDCIGPETQSFWLRPHAQGFRDL LNWLSKRYGYPKIYVTENGTSLKGENDMPLEQVLEDDFRVKYFNDYVRAMAAAVAEDGCN VRGYLAWSLLDNFEWAEGYETRFGVTYVDYANDQKRYPKKSAKSLKPLFDSLIRKE |
Below you can see a comparison between both sequences generated using the Clustal Omega web tool (available at https://www.ebi.ac.uk/Tools/msa/clustalo/). Humicola insolens β-glucosidase is shown above, and Uncultured bacteria's β-glucosidase sequence from the South China Sea is shown below. The asterisks (*) represent conserved amino acid residues, while the punctuation mark (:) represents a modification for an amino acid residue with similar characteristics. Take note that β-glucosidase sequences from the same family present similar sequence.
Humicola ----------MSLPPDFKWGFATAAYQIEGSVNEDGRGPSIWDTFCAIPGKIADGSSGAV 50 uncultured MTKISLPTCSPLLTKEFIYGVATASFQIEGGS--AHRLPCIWDTFCDTPGKIADNSNGHV 58 * :* :*.***::****. * *.****** ******.*.* * Humicola ACDSYKRTKEDIALLKELGANSYRFSISWSRIIPLGGRNDPINQKGIDHYVKFVDDLIEA 110 uncultured ACDHYNNWKQDIDLIESLGVDAYRLSISWPRVITKSGE---LNPEGVKFYTDILDELKKR 115 *** *:. *:** *::.**.::**:**** *:* .*. :* :*:..*..::*:* : Humicola GITPFITLFHWDLPDALDKRYGGFLNKEEFAADFENYARIM-FKAIPKCKHWITFNEPWC 169 uncultured NIKAFVTLYHWDLPQHLEDE-GGWLNRETA-YAFAHYVDLITLAFGDRVHSYATLNEPFC 173 .*. *:**:*****: *:.. **:**:* * :*. :: : : : : *:***:* Humicola SAILGYNTGYFAPGHTSDRSKSPVGDSAREPWIVGHNILIAHARAVKAYREDFKPTQGGE 229 uncultured SAFLGYEIGIHAPGKVGKQYGR----K------AAHHLLLAHGLAMTVLKQNSPTTLN-- 221 **:***: * .***:...: . ..*::*:**. *:.. ::: * . Humicola IGITLNGDATLPWDPEDPADIEACDRKIEFAISWFADPIYFGKYPDSMRKQLGDRLPEFT 289 uncultured -GIVLNFTPCY-SISEDADDIAATAFADDYLNQWYMKPIMDGTYPAIIEQLPSAHLPDIH 279 **.** ** ** * :: .*: .** *.** :.: . :**:: Humicola PEEVALVKGSNDFYGMNHYTANYIKHKTGVPPEDDFLGNLETLFYNKYGDCIGPETQSFW 349 uncultured DGDMAIISQSIDYLGINFYTRQFYKAHPTEI-------------YEP-IEPTGPLTDMGW 325 ::*::. * *: *:*.** :: * : *: : ** *: * Humicola LRPHAQGFRDLLNWLSKRYGYPKIYVTENGTSLKGENDMPLEQVLEDDFRVKYFNDYVRA 409 uncultured E-IYPKSFTELLVTLNNTYTLPPIFITENGAAMPDSYNN---GEINDVDRLDYYNSHLNA 381 : :.* :** *.: * * *::****::: .. : ::* *:.*:*.::.* Humicola MAAAVAEDGCNVRGYLAWSLLDNFEWAEGYETRFGVTYVDYANDQKRYPKKSAKSLKPLF 469 uncultured VH-NATEQGVRIDGYFAWSLMDNFEWAEGYLKRFGIVYVDYSTQQRTIKNSG-LAYKALI 439 : .:*:* .: **:****:********* .***:.****:.:*: :.. : * *: Humicola DSLIRKE 476 uncultured ----SNR 442 :. |
Also, proteins from the GH1 family have a conserved folding structure called TIM-barrel [7]. Figure 1 illustrates the three-dimensional structure of GH1 β-glucosidase from the fungus Humicola insolens complexed with a glucose molecule [8].
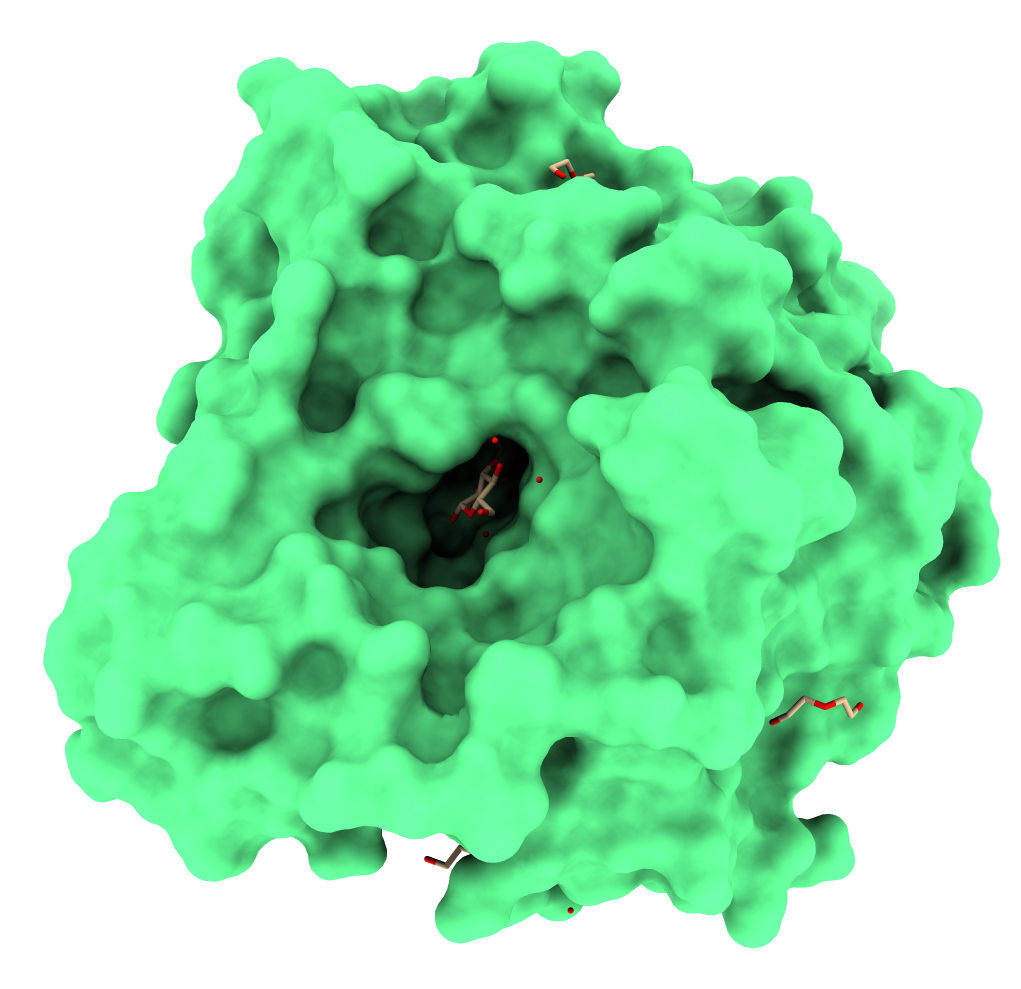 Figure 1. GH1 β-glucosidase from the fungus Humicola insolens. Obtained from PDB ID: 4MDP. Figure generated using ChimeraX [9].
Figure 1. GH1 β-glucosidase from the fungus Humicola insolens. Obtained from PDB ID: 4MDP. Figure generated using ChimeraX [9].
In conclusion, β-glucosidase enzymes have excellent potential for second-generation biofuel production. However, genetic engineering applications are still necessary to improve the activity of non-tolerant enzymes. Additionally, bioinformatics applications have been successfully used to bring new insights to detect sites for mutations that could improve their activity.
References
- James R. Ketudat Cairns; Asim Esen; β-Glucosidases. Cellular and Molecular Life Sciences 2010, 67, 3389-3405, 10.1007/s00018-010-0399-2.
- Leon Sulfierry Corrêa Costa; Diego César Batista Mariano; Rafael Eduardo Oliveira Rocha; Johannes Kraml; Carlos Henrique Da Silveira; Klaus Roman Liedl; Raquel Cardoso De Melo-Minardi; Leonardo Henrique Franca De Lima; Molecular Dynamics Gives New Insights into the Glucose Tolerance and Inhibition Mechanisms on β-Glucosidases.. Molecules 2019, 24, 3215, 10.3390/molecules24183215.
- Diego Mariano; Naiara Pantuza; Lucianna H. Santos; Rafael E. O. Rocha; Leonardo Lima; Lucas Bleicher; Raquel Cardoso De Melo-Minardi; Glutantβase: a database for improving the rational design of glucose-tolerant β-glucosidases. BMC Molecular and Cell Biology 2020, 21, 1-15, 10.1186/s12860-020-00293-y.
- Andreza P. Garbin; Nayara F.L. Garcia; Gabriela F. Cavalheiro; Maria Alice Silvestre; André Rodrigues; Marcelo F. DA Paz; Gustavo G. Fonseca; Rodrigo S.R. Leite; β-glucosidase from thermophilic fungus Thermoascus crustaceus: production and industrial potential. Anais da Academia Brasileira de Ciências 2020, 93, e20191349, 10.1590/0001-3765202120191349.
- Diego Mariano; C. Leite; L.H.S. Santos; L.F. Marins; K.S. Machado; Adriano Velasque Werhli; L.H.F. Lima; R.C. De Melo-Minardi; Characterization of glucose-tolerant β-glucosidases used in biofuel production under the bioinformatics perspective: a systematic review. Genetics and Molecular Research 2013, 16, 1, 10.4238/gmr16039740.
- Diego César Batista Mariano; Lucianna Helene Santos; Karina Dos Santos Machado; Adriano Velasque Werhli; Leonardo Henrique França De Lima; Raquel Cardoso De Melo-Minardi; A Computational Method to Propose Mutations in Enzymes Based on Structural Signature Variation (SSV). International Journal of Molecular Sciences 2019, 20, 333, 10.3390/ijms20020333.
- Nozomi Nagano; Christine A Orengo; Janet Thornton; One Fold with Many Functions: The Evolutionary Relationships between TIM Barrel Families Based on their Sequences, Structures and Functions. Journal of Molecular Biology 2002, 321, 741-765, 10.1016/s0022-2836(02)00649-6.
- Priscila Oliveira de Giuseppe; Tatiana De Arruda Campos Brasil Souza; Flavio Henrique Moreira Souza; Leticia Maria Zanphorlin; Carla Botelho Machado; Richard Ward; Joao Atilio Jorge; Rosa Dos Prazeres Melo Furriel; Mario Tyago Murakami; Structural basis for glucose tolerance in GH1 β-glucosidases. Acta Crystallographica Section D Biological Crystallography 2014, 70, 1631-1639, 10.1107/s1399004714006920.
- Eric F. Pettersen; Thomas D. Goddard; Conrad C. Huang; Elaine C. Meng; Gregory S. Couch; Tristan I. Croll; John H. Morris; Thomas E. Ferrin; UCSF ChimeraX : Structure visualization for researchers, educators, and developers. Protein Science 2020, 30, 70-82, 10.1002/pro.3943.
