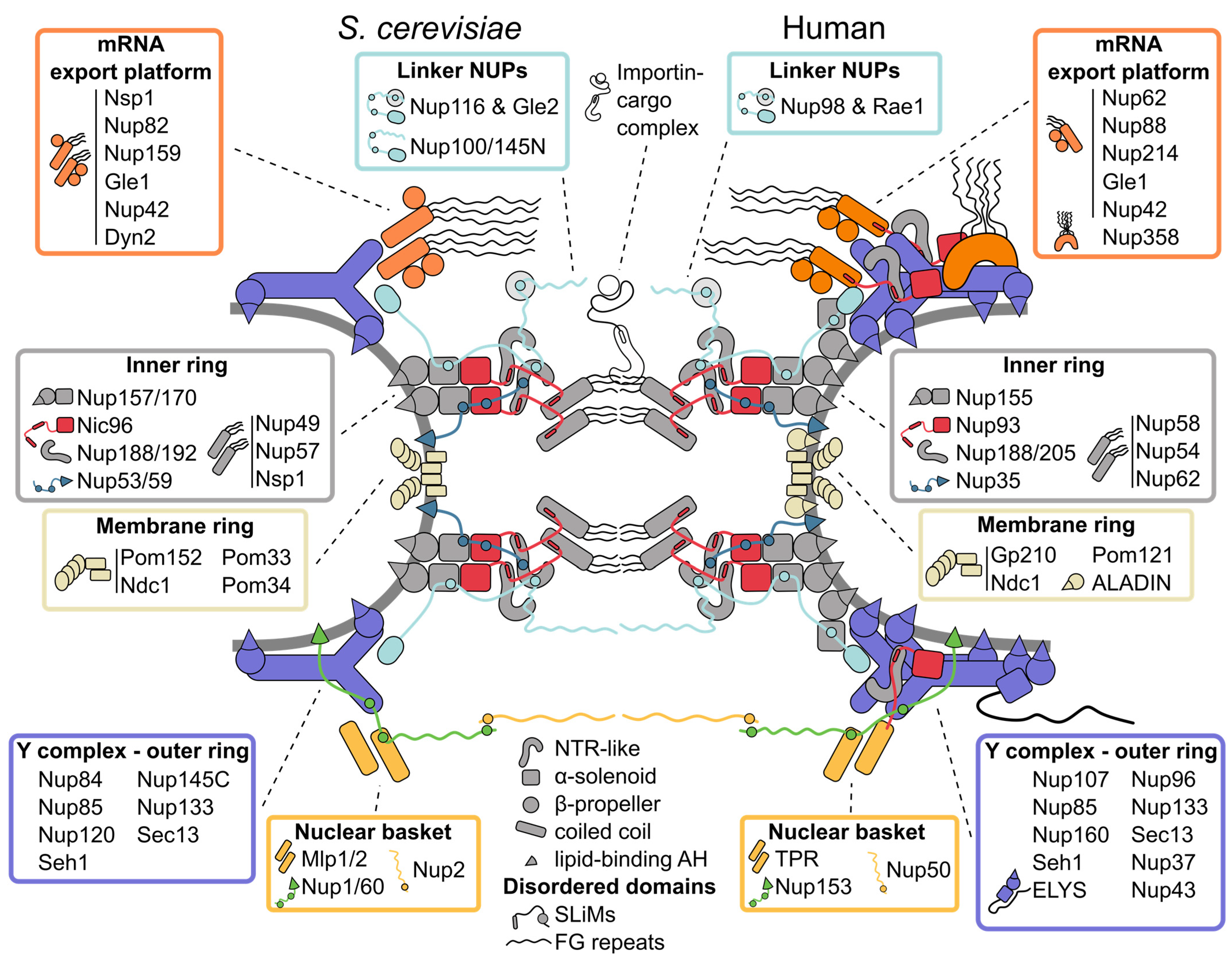
Nuclear pore complexes (NPCs) are the only transport channels that cross the nuclear envelope. Constructed from ~500–1000 nucleoporin proteins each, they are among the largest macromolecular assemblies in eukaryotic cells. Thanks to advances in structural analysis approaches, the construction principles and architecture of the NPC have recently been revealed at submolecular resolution. Although the overall structure and inventory of nucleoporins are conserved, NPCs exhibit significant compositional and functional plasticity even within single cells and surprising variability in their assembly pathways. Once assembled, NPCs remain seemingly unexchangeable in post-mitotic cells.
- nuclear pore complex
- nucleoporin
- NPC
1. Inner Ring: The Flexible Core of the Nuclear Pore Complex
2. Symmetric Outer Rings: The Versatile Outer Coat of the Nuclear Pore Complex
3. Asymmetric Appendages: Functional Extensions of the Nuclear Pore Complex
4. The Membrane Ring: An Enigmatic Girdle
5. Linker Nucleoporins: An Invisible Thread Stitching the Nuclear Pore Complex Together
This entry is adapted from the peer-reviewed paper 10.3390/cells11091456
References
- DeGrasse, J.A.; DuBois, K.N.; Devos, D.; Siegel, T.N.; Sali, A.; Field, M.C.; Rout, M.P.; Chait, B.T. Evidence for a shared nuclear pore complex architecture that is conserved from the last common eukaryotic ancestor. Mol. Cell Proteom. 2009, 8, 2119–2130.
- Stuwe, T.; Bley, C.J.; Thierbach, K.; Petrovic, S.; Schilbach, S.; Mayo, D.J.; Perriches, T.; Rundlet, E.J.; Jeon, Y.E.; Collins, L.N.; et al. Architecture of the fungal nuclear pore inner ring complex. Science 2015, 350, 56–64.
- Whittle, J.R.R.; Schwartz, T.U. Architectural nucleoporins Nup157/170 and Nup133 are structurally related and descend from a second ancestral element. J. Biol. Chem. 2009, 284, 28442–28452.
- Lin, D.H.; Stuwe, T.; Schilbach, S.; Rundlet, E.J.; Perriches, T.; Mobbs, G.; Fan, Y.; Thierbach, K.; Huber, F.M.; Collins, L.N.; et al. Architecture of the symmetric core of the nuclear pore. Science 2016, 352, aaf1015.
- Andersen, K.R.; Onischenko, E.; Tang, J.H.; Kumar, P.; Chen, J.Z.; Ulrich, A.; Liphardt, J.T.; Weis, K.; Schwartz, T.U. Scaffold nucleoporins Nup188 and Nup192 share structural and functional properties with nuclear transport receptors. eLife 2013, 2, e00745.
- Flemming, D.; Devos, D.P.; Schwarz, J.; Amlacher, S.; Lutzmann, M.; Hurt, E. Analysis of the yeast nucleoporin Nup188 reveals a conserved S-like structure with similarity to karyopherins. J. Struct. Biol. 2012, 177, 99–105.
- Stuwe, T.; Lin, D.H.; Collins, L.N.; Hurt, E.; Hoelz, A. Evidence for an evolutionary relationship between the large adaptor nucleoporin Nup192 and karyopherins. Proc. Natl. Acad. Sci. USA 2014, 111, 2530–2535.
- Akey, C.W.; Singh, D.; Ouch, C.; Echeverria, I.; Nudelman, I.; Varberg, J.M.; Yu, Z.; Fang, F.; Shi, Y.; Wang, J.; et al. Comprehensive structure and functional adaptations of the yeast nuclear pore complex. Cell 2022, 185, 361–378.e25.
- Li, Z.; Chen, S.; Zhao, L.; Huang, G.; Pi, X.; Sun, S.; Wang, P.; Sui, S.F. Near-atomic structure of the inner ring of the Saccharomyces cerevisiae nuclear pore complex. Cell Res. 2022, 1–14.
- Petrovic, S.; Samanta, D.; Perriches, T.; Bley, C.J.; Thierbach, K.; Brown, B.; Nie, S.; Mobbs, G.W.; Stevens, T.A.; Liu, X.; et al. Architecture of the linker-scaffold in the nuclear pore. bioRxiv 2021.
- Bley, C.J.; Nie, S.; Mobbs, G.W.; Petrovic, S.; Gres, A.T.; Liu, X.; Mukherjee, S.; Harvey, S.; Huber, F.M.; Lin, D.H.; et al. Architecture of the cytoplasmic face of the nuclear pore. bioRxiv 2021.
- Fischer, J.; Teimer, R.; Amlacher, S.; Kunze, R.; Hurt, E. Linker Nups connect the nuclear pore complex inner ring with the outer ring and transport channel. Nat. Struct. Mol. Biol. 2015, 22, 774–781.
- Onischenko, E.; Stanton, L.H.; Madrid, A.S.; Kieselbach, T.; Weis, K. Role of the Ndc1 interaction network in yeast nuclear pore complex assembly and maintenance. J. Cell Biol. 2009, 185, 475–491.
- Mosalaganti, S.; Obarska-Kosinska, A.; Siggel, M.; Turonova, B.; Zimmerli, C.E.; Buczak, K.; Schmidt, F.H.; Margiotta, E.; Mackmull, M.-T.; Hagen, W.; et al. Artificial intelligence reveals nuclear pore complexity. bioRxiv 2021.
- Schuller, A.P.; Wojtynek, M.; Mankus, D.; Tatli, M.; Kronenberg-Tenga, R.; Regmi, S.G.; Dip, P.V.; Lytton-Jean, A.K.R.; Brignole, E.J.; Dasso, M.; et al. The cellular environment shapes the nuclear pore complex architecture. Nature 2021, 598, 667–671.
- Zimmerli, C.E.; Allegretti, M.; Rantos, V.; Goetz, S.K.; Obarska-Kosinska, A.; Zagoriy, I.; Halavatyi, A.; Hummer, G.; Mahamid, J.; Kosinski, J.; et al. Nuclear pores dilate and constrict in cellulo. Science 2021, 374, eabd9776.
- Zila, V.; Margiotta, E.; Turonova, B.; Muller, T.G.; Zimmerli, C.E.; Mattei, S.; Allegretti, M.; Borner, K.; Rada, J.; Muller, B.; et al. Cone-shaped HIV-1 capsids are transported through intact nuclear pores. Cell 2021, 184, 1032–1046.e18.
- Liashkovich, I.; Meyring, A.; Kramer, A.; Shahin, V. Exceptional structural and mechanical flexibility of the nuclear pore complex. J. Cell Physiol. 2011, 226, 675–682.
- Tai, L.; Zhu, Y.; Ren, H.; Huang, X.; Zhang, C.; Sun, F. 8 Å structure of the outer rings of the Xenopus laevis nuclear pore complex obtained by cryo-EM and AI. Protein Cell 2022, 1–18.
- Sampathkumar, P.; Kim, S.J.; Upla, P.; Rice, W.J.; Phillips, J.; Timney, B.L.; Pieper, U.; Bonanno, J.B.; Fernandez-Martinez, J.; Hakhverdyan, Z.; et al. Structure, dynamics, evolution, and function of a major scaffold component in the nuclear pore complex. Structure 2013, 21, 560–571.
- Bui, K.H.; von Appen, A.; DiGuilio, A.L.; Ori, A.; Sparks, L.; Mackmull, M.T.; Bock, T.; Hagen, W.; Andres-Pons, A.; Glavy, J.S.; et al. Integrated structural analysis of the human nuclear pore complex scaffold. Cell 2013, 155, 1233–1243.
- Maimon, T.; Elad, N.; Dahan, I.; Medalia, O. The human nuclear pore complex as revealed by cryo-electron tomography. Structure 2012, 20, 998–1006.
- Kelley, K.; Knockenhauer, K.E.; Kabachinski, G.; Schwartz, T.U. Atomic structure of the Y complex of the nuclear pore. Nat. Struct. Mol. Biol. 2015, 22, 425–431.
- Thierbach, K.; von Appen, A.; Thoms, M.; Beck, M.; Flemming, D.; Hurt, E. Protein interfaces of the conserved Nup84 complex from Chaetomium thermophilum shown by crosslinking mass spectrometry and electron microscopy. Structure 2013, 21, 1672–1682.
- Liu, H.L.; De Souza, C.P.; Osmani, A.H.; Osmani, S.A. The three fungal transmembrane nuclear pore complex proteins of Aspergillus nidulans are dispensable in the presence of an intact An-Nup84-120 complex. Mol. Biol. Cell 2009, 20, 616–630.
- Kampmann, M.; Blobel, G. Three-dimensional structure and flexibility of a membrane-coating module of the nuclear pore complex. Nat. Struct. Mol. Biol. 2009, 16, 782–788.
- Lutzmann, M.; Kunze, R.; Buerer, A.; Aebi, U.; Hurt, E. Modular self-assembly of a Y-shaped multiprotein complex from seven nucleoporins. EMBO J. 2002, 21, 387–397.
- Nordeen, S.A.; Turman, D.L.; Schwartz, T.U. Yeast Nup84-Nup133 complex structure details flexibility and reveals conservation of the membrane anchoring ALPS motif. Nat. Commun. 2020, 11, 6060.
- Siniossoglou, S.; Lutzmann, M.; Santos-Rosa, H.; Leonard, K.; Mueller, S.; Aebi, U.; Hurt, E. Structure and assembly of the Nup84p complex. J. Cell Biol. 2000, 149, 41–54.
- Allegretti, M.; Zimmerli, C.E.; Rantos, V.; Wilfling, F.; Ronchi, P.; Fung, H.K.H.; Lee, C.W.; Hagen, W.; Turonova, B.; Karius, K.; et al. In-cell architecture of the nuclear pore and snapshots of its turnover. Nature 2020, 586, 796–800.
- Kim, S.J.; Fernandez-Martinez, J.; Nudelman, I.; Shi, Y.; Zhang, W.; Raveh, B.; Herricks, T.; Slaughter, B.D.; Hogan, J.A.; Upla, P.; et al. Integrative structure and functional anatomy of a nuclear pore complex. Nature 2018, 555, 475–482.
- von Appen, A.; Kosinski, J.; Sparks, L.; Ori, A.; DiGuilio, A.L.; Vollmer, B.; Mackmull, M.T.; Banterle, N.; Parca, L.; Kastritis, P.; et al. In situ structural analysis of the human nuclear pore complex. Nature 2015, 526, 140–143.
- Eibauer, M.; Pellanda, M.; Turgay, Y.; Dubrovsky, A.; Wild, A.; Medalia, O. Structure and gating of the nuclear pore complex. Nat. Commun. 2015, 6, 7532.
- Huang, G.; Zhang, Y.; Zhu, X.; Zeng, C.; Wang, Q.; Zhou, Q.; Tao, Q.; Liu, M.; Lei, J.; Yan, C.; et al. Structure of the cytoplasmic ring of the Xenopus laevis nuclear pore complex by cryo-electron microscopy single particle analysis. Cell Res. 2020, 30, 520–531.
- Mosalaganti, S.; Kosinski, J.; Albert, S.; Schaffer, M.; Strenkert, D.; Salome, P.A.; Merchant, S.S.; Plitzko, J.M.; Baumeister, W.; Engel, B.D.; et al. In situ architecture of the algal nuclear pore complex. Nat. Commun. 2018, 9, 2361.
- Berke, I.C.; Boehmer, T.; Blobel, G.; Schwartz, T.U. Structural and functional analysis of Nup133 domains reveals modular building blocks of the nuclear pore complex. J. Cell Biol. 2004, 167, 591–597.
- Drin, G.; Casella, J.F.; Gautier, R.; Boehmer, T.; Schwartz, T.U.; Antonny, B. A general amphipathic alpha-helical motif for sensing membrane curvature. Nat. Struct. Mol. Biol. 2007, 14, 138–146.
- Leksa, N.C.; Brohawn, S.G.; Schwartz, T.U. The structure of the scaffold nucleoporin Nup120 reveals a new and unexpected domain architecture. Structure 2009, 17, 1082–1091.
- Brohawn, S.G.; Leksa, N.C.; Spear, E.D.; Rajashankar, K.R.; Schwartz, T.U. Structural evidence for common ancestry of the nuclear pore complex and vesicle coats. Science 2008, 322, 1369–1373.
- Debler, E.W.; Ma, Y.; Seo, H.S.; Hsia, K.C.; Noriega, T.R.; Blobel, G.; Hoelz, A. A fence-like coat for the nuclear pore membrane. Mol. Cell 2008, 32, 815–826.
- Hsia, K.C.; Stavropoulos, P.; Blobel, G.; Hoelz, A. Architecture of a coat for the nuclear pore membrane. Cell 2007, 131, 1313–1326.
- Field, M.C.; Rout, M.P. Pore timing: The evolutionary origins of the nucleus and nuclear pore complex. F1000Res 2019, 8.
- Hinshaw, J.E.; Milligan, R.A. Nuclear pore complexes exceeding eightfold rotational symmetry. J. Struct. Biol. 2003, 141, 259–268.
- Loschberger, A.; Franke, C.; Krohne, G.; van de Linde, S.; Sauer, M. Correlative super-resolution fluorescence and electron microscopy of the nuclear pore complex with molecular resolution. J. Cell Sci. 2014, 127, 4351–4355.
- Onischenko, E.; Tang, J.H.; Andersen, K.R.; Knockenhauer, K.E.; Vallotton, P.; Derrer, C.P.; Kralt, A.; Mugler, C.F.; Chan, L.Y.; Schwartz, T.U.; et al. Natively Unfolded FG Repeats Stabilize the Structure of the Nuclear Pore Complex. Cell 2017, 171, 904–917.e19.
- Franz, C.; Walczak, R.; Yavuz, S.; Santarella, R.; Gentzel, M.; Askjaer, P.; Galy, V.; Hetzer, M.; Mattaj, I.W.; Antonin, W. MEL-28/ELYS is required for the recruitment of nucleoporins to chromatin and postmitotic nuclear pore complex assembly. EMBO Rep. 2007, 8, 165–172.
- Rasala, B.A.; Orjalo, A.V.; Shen, Z.; Briggs, S.; Forbes, D.J. ELYS is a dual nucleoporin/kinetochore protein required for nuclear pore assembly and proper cell division. Proc. Natl. Acad. Sci. USA 2006, 103, 17801–17806.
- Rasala, B.A.; Ramos, C.; Harel, A.; Forbes, D.J. Capture of AT-rich chromatin by ELYS recruits POM121 and NDC1 to initiate nuclear pore assembly. Mol. Biol. Cell 2008, 19, 3982–3996.
- Jarnik, M.; Aebi, U. Toward a more complete 3-D structure of the nuclear pore complex. J. Struct. Biol. 1991, 107, 291–308.
- Goldberg, M.W.; Allen, T.D. The nuclear pore complex: Three-dimensional surface structure revealed by field emission, in-lens scanning electron microscopy, with underlying structure uncovered by proteolysis. J. Cell Sci. 1993, 106 Pt 1, 261–274.
- Watson, M.L. Further observations on the nuclear envelope of the animal cell. J. Biophys Biochem. Cytol. 1959, 6, 147–156.
- Fernandez-Martinez, J.; Kim, S.J.; Shi, Y.; Upla, P.; Pellarin, R.; Gagnon, M.; Chemmama, I.E.; Wang, J.; Nudelman, I.; Zhang, W.; et al. Structure and Function of the Nuclear Pore Complex Cytoplasmic mRNA Export Platform. Cell 2016, 167, 1215–1228.e25.
- Walther, T.C.; Pickersgill, H.S.; Cordes, V.C.; Goldberg, M.W.; Allen, T.D.; Mattaj, I.W.; Fornerod, M. The cytoplasmic filaments of the nuclear pore complex are dispensable for selective nuclear protein import. J. Cell Biol. 2002, 158, 63–77.
- Vallotton, P.; Rajoo, S.; Wojtynek, M.; Onischenko, E.; Kralt, A.; Derrer, C.P.; Weis, K. Mapping the native organization of the yeast nuclear pore complex using nuclear radial intensity measurements. Proc. Natl. Acad. Sci. USA 2019, 116, 14606–14613.
- Lin, D.H.; Hoelz, A. The Structure of the Nuclear Pore Complex (An Update). Annu. Rev. Biochem. 2019, 88, 725–783.
- Obado, S.O.; Brillantes, M.; Uryu, K.; Zhang, W.; Ketaren, N.E.; Chait, B.T.; Field, M.C.; Rout, M.P. Interactome Mapping Reveals the Evolutionary History of the Nuclear Pore Complex. PLoS Biol. 2016, 14, e1002365.
- Goldberg, M.W.; Allen, T.D. High resolution scanning electron microscopy of the nuclear envelope: Demonstration of a new, regular, fibrous lattice attached to the baskets of the nucleoplasmic face of the nuclear pores. J. Cell Biol. 1992, 119, 1429–1440.
- Kiseleva, E.; Allen, T.D.; Rutherford, S.; Bucci, M.; Wente, S.R.; Goldberg, M.W. Yeast nuclear pore complexes have a cytoplasmic ring and internal filaments. J. Struct. Biol. 2004, 145, 272–288.
- Krull, S.; Thyberg, J.; Bjorkroth, B.; Rackwitz, H.R.; Cordes, V.C. Nucleoporins as components of the nuclear pore complex core structure and Tpr as the architectural element of the nuclear basket. Mol. Biol. Cell 2004, 15, 4261–4277.
- Niepel, M.; Molloy, K.R.; Williams, R.; Farr, J.C.; Meinema, A.C.; Vecchietti, N.; Cristea, I.M.; Chait, B.T.; Rout, M.P.; Strambio-De-Castillia, C. The nuclear basket proteins Mlp1p and Mlp2p are part of a dynamic interactome including Esc1p and the proteasome. Mol. Biol. Cell 2013, 24, 3920–3938.
- Li, Y.; Aksenova, V.; Tingey, M.; Yu, J.; Ma, P.; Arnaoutov, A.; Chen, S.; Dasso, M.; Yang, W. Distinct roles of nuclear basket proteins in directing the passage of mRNA through the nuclear pore. Proc. Natl. Acad. Sci. USA 2021, 118.
- Mi, L.; Goryaynov, A.; Lindquist, A.; Rexach, M.; Yang, W. Quantifying nucleoporin stoichiometry inside single nuclear pore complexes in vivo. Sci. Rep. 2015, 5, 9372.
- Ori, A.; Banterle, N.; Iskar, M.; Andres-Pons, A.; Escher, C.; Khanh Bui, H.; Sparks, L.; Solis-Mezarino, V.; Rinner, O.; Bork, P.; et al. Cell type-specific nuclear pores: A case in point for context-dependent stoichiometry of molecular machines. Mol. Syst. Biol. 2013, 9, 648.
- Rajoo, S.; Vallotton, P.; Onischenko, E.; Weis, K. Stoichiometry and compositional plasticity of the yeast nuclear pore complex revealed by quantitative fluorescence microscopy. Proc. Natl. Acad. Sci. USA 2018, 115, E3969–E3977.
- Albert, S.; Schaffer, M.; Beck, F.; Mosalaganti, S.; Asano, S.; Thomas, H.F.; Plitzko, J.M.; Beck, M.; Baumeister, W.; Engel, B.D. Proteasomes tether to two distinct sites at the nuclear pore complex. Proc. Natl. Acad. Sci. USA 2017, 114, 13726–13731.
- Cibulka, J.; Bisaccia, F.; Radisavljevic, K.; Gudino Carrillo, R.M.; Kohler, A. Assembly principle of a membrane-anchored nuclear pore basket scaffold. Sci. Adv. 2022, 8, eabl6863.
- Mészáros, N.; Cibulka, J.; Mendiburo, M.J.; Romanauska, A.; Schneider, M.; Köhler, A. Nuclear pore basket proteins are tethered to the nuclear envelope and can regulate membrane curvature. Dev. Cell 2015, 33, 285–298.
- Vollmer, B.; Lorenz, M.; Moreno-Andres, D.; Bodenhofer, M.; De Magistris, P.; Astrinidis, S.A.; Schooley, A.; Flotenmeyer, M.; Leptihn, S.; Antonin, W. Nup153 Recruits the Nup107-160 Complex to the Inner Nuclear Membrane for Interphasic Nuclear Pore Complex Assembly. Dev. Cell 2015, 33, 717–728.
- Hase, M.E.; Cordes, V.C. Direct interaction with nup153 mediates binding of Tpr to the periphery of the nuclear pore complex. Mol. Biol. Cell 2003, 14, 1923–1940.
- Makise, M.; Mackay, D.R.; Elgort, S.; Shankaran, S.S.; Adam, S.A.; Ullman, K.S. The Nup153-Nup50 protein interface and its role in nuclear import. J. Biol. Chem. 2012, 287, 38515–38522.
- Bensidoun, P.; Zenklusen, D.; Oeffinger, M. Choosing the right exit: How functional plasticity of the nuclear pore drives selective and efficient mRNA export. Wiley Interdiscip. Rev. RNA 2021, 12, e1660.
- Neumann, N.; Lundin, D.; Poole, A.M. Comparative genomic evidence for a complete nuclear pore complex in the last eukaryotic common ancestor. PLoS ONE 2010, 5, e13241.
- Field, M.C.; Koreny, L.; Rout, M.P. Enriching the pore: Splendid complexity from humble origins. Traffic 2014, 15, 141–156.
- Chadrin, A.; Hess, B.; San Roman, M.; Gatti, X.; Lombard, B.; Loew, D.; Barral, Y.; Palancade, B.; Doye, V. Pom33, a novel transmembrane nucleoporin required for proper nuclear pore complex distribution. J. Cell Biol. 2010, 189, 795–811.
- Stavru, F.; Hulsmann, B.B.; Spang, A.; Hartmann, E.; Cordes, V.C.; Gorlich, D. NDC1: A crucial membrane-integral nucleoporin of metazoan nuclear pore complexes. J. Cell Biol. 2006, 173, 509–519.
- Tcheperegine, S.E.; Marelli, M.; Wozniak, R.W. Topology and functional domains of the yeast pore membrane protein Pom152p. J. Biol. Chem. 1999, 274, 5252–5258.
- Upla, P.; Kim, S.J.; Sampathkumar, P.; Dutta, K.; Cahill, S.M.; Chemmama, I.E.; Williams, R.; Bonanno, J.B.; Rice, W.J.; Stokes, D.L.; et al. Molecular Architecture of the Major Membrane Ring Component of the Nuclear Pore Complex. Structure 2017, 25, 434–445.
- Hao, Q.; Zhang, B.; Yuan, K.; Shi, H.; Blobel, G. Electron microscopy of Chaetomium pom152 shows the assembly of ten-bead string. Cell Discov. 2018, 4, 56.
- Zhang, Y.; Li, S.; Zeng, C.; Huang, G.; Zhu, X.; Wang, Q.; Wang, K.; Zhou, Q.; Yan, C.; Zhang, W.; et al. Molecular architecture of the luminal ring of the Xenopus laevis nuclear pore complex. Cell Res. 2020, 30, 532–540.
- Stavru, F.; Nautrup-Pedersen, G.; Cordes, V.C.; Gorlich, D. Nuclear pore complex assembly and maintenance in POM121- and gp210-deficient cells. J. Cell Biol. 2006, 173, 477–483.
- Wozniak, R.W.; Blobel, G.; Rout, M.P. POM152 is an integral protein of the pore membrane domain of the yeast nuclear envelope. J. Cell Biol. 1994, 125, 31–42.
- Olsson, M.; Scheele, S.; Ekblom, P. Limited expression of nuclear pore membrane glycoprotein 210 in cell lines and tissues suggests cell-type specific nuclear pores in metazoans. Exp. Cell Res. 2004, 292, 359–370.
- D’Angelo, M.A.; Raices, M.; Panowski, S.H.; Hetzer, M.W. Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell 2009, 136, 284–295.
- Rabut, G.; Doye, V.; Ellenberg, J. Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat. Cell Biol. 2004, 6, 1114–1121.
- Savas, J.N.; Toyama, B.H.; Xu, T.; Yates, J.R., 3rd; Hetzer, M.W. Extremely long-lived nuclear pore proteins in the rat brain. Science 2012, 335, 942.
- Toyama, B.H.; Arrojo, E.D.R.; Lev-Ram, V.; Ramachandra, R.; Deerinck, T.J.; Lechene, C.; Ellisman, M.H.; Hetzer, M.W. Visualization of long-lived proteins reveals age mosaicism within nuclei of postmitotic cells. J. Cell Biol. 2019, 218, 433–444.
- Toyama, B.H.; Savas, J.N.; Park, S.K.; Harris, M.S.; Ingolia, N.T.; Yates, J.R., 3rd; Hetzer, M.W. Identification of long-lived proteins reveals exceptional stability of essential cellular structures. Cell 2013, 154, 971–982.
- Knockenhauer, K.E.; Schwartz, T.U. The nuclear pore complex as a flexible and dynamic gate. Cell 2016, 164, 1162–1171.
- Dultz, E.; Zanin, E.; Wurzenberger, C.; Braun, M.; Rabut, G.; Sironi, L.; Ellenberg, J. Systematic kinetic analysis of mitotic dis- and reassembly of the nuclear pore in living cells. J. Cell Biol. 2008, 180, 857–865.
- Amlacher, S.; Sarges, P.; Flemming, D.; van Noort, V.; Kunze, R.; Devos, D.P.; Arumugam, M.; Bork, P.; Hurt, E. Insight into structure and assembly of the nuclear pore complex by utilizing the genome of a eukaryotic thermophile. Cell 2011, 146, 277–289.
- Teimer, R.; Kosinski, J.; von Appen, A.; Beck, M.; Hurt, E. A short linear motif in scaffold Nup145C connects Y-complex with pre-assembled outer ring Nup82 complex. Nat. Commun. 2017, 8, 1107.
- Hamed, M.; Antonin, W. Dunking into the Lipid Bilayer: How Direct Membrane Binding of Nucleoporins Can Contribute to Nuclear Pore Complex Structure and Assembly. Cells 2021, 10, 3601.
