You're using an outdated browser. Please upgrade to a modern browser for the best experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Plant pathogen presence is very dangerous for agricultural ecosystems and causes huge economic losses. Phytoplasmas are insect-transmitted wall-less bacteria living in plants, only in the phloem tissues and in the emolymph of their insect vectors. They are able to manipulate several metabolic pathways of their hosts, very often without impairing their life. The molecular diversity described (49 ‘Candidatus Phytoplasma’ species and about 300 ribosomal subgroups) is only in some cases related to their associated symptomatology.
- plant diseases
- pathogenicity
- molecular classification
1. Introduction
Plant pathogen presence is usually known as a very dangerous component of agricultural ecosystems and is associated with huge economic losses. The world history was also often shaped by dangerous plant epidemics or pandemics such as the wheat rust that was among the main causes of the Roman empire failure, the potato late blight by Phythophthora infestans producing the Irish migration to America due to the famine, and the coffee rust obliging to stop the coffee cultivation is several areas, mainly in islands. Recently plant pathogenic bacteria have played an important role in reducing kiwi cultivation, due to the canker by Pseudomonas syringae pv. actinidiae [1], and citrus, through the greening (‘Candidatus Liberibacter’ species) [2] diseases. Moreover, there are bacteria hosted by plants and insects that are both associated with severe epidemic or with useful changes in plant behavior. While their presence in apple trees causes severe losses in production and kills millions of coconut palm trees in the Caribbean, the presence of a poinsettia branching bacterium is allowing its commercial production as pot plant (Figure 1).

Figure 1. Coconut (a), aster (b), and poinsettia (c) infected by phytoplasmas.
Phytoplasmas are insect-transmitted wall-less bacteria provisionally classified to be the ‘Candidatus Phytoplasma’ species [3][4]. They live only in the plant phloem tissues and in the emolymph of their insect vectors, especially concentrated in the salivary glands. Their relationship with both plants and insects is very intimate and they are able to manipulate several metabolic pathways, very often without impairing the host’s life [5].
2. Phytoplasma Discovery
The phytoplasma presence in plants historically dates back about 1000 years, when special tree peonies exhibiting green flowers were given to the Chinese court during the Song dynasty (900 BC) as the most precious and beautiful flower of the empire. However, scientific records of phytoplasma-associated plant diseases started when, in 1967, mulberry dwarf, rice yellow dwarf, and sweet potato witches’ broom, long considered to be caused by viruses, using electron microscopy, were found to be colonized by small pleomorphic bodies (80–800 nm in diameter) resembling mycoplasmas (bacterial pathogens of humans and animals) and were named mycoplasma-like organisms (MLOs) [6]. Their discovery stimulated worldwide investigation and numerous plant diseases were associated with the consistent presence of MLOs. These bacteria were long considered unculturable, but about 10 years ago, colonies containing molecularly different phytoplasmas began to be obtained in artificial media from different infected plant species (Figure 2) [7][8][9][10][11].
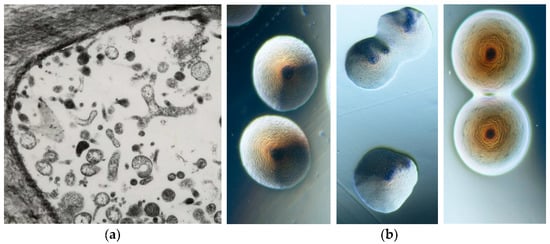
Figure 2. Transmission electron microscopy picture of a thin section in the phloematic tissue of a phytoplasma infected gladiolus plant showing the presence of strong pleomorphism (×8000) (a). Tree morphotypes of colonies containing phytoplasmas under binocular microscope (×40) (b).
3. Phytoplasma Classification
The ‘Candidatus Phytoplasma’ genus provisional classification is highly relevant due to its application in epidemiological and ecological studies, mainly aimed at keeping the severe phytoplasma plant diseases under control worldwide. The updated proposed guidelines accommodate those ‘Ca. Phytoplasma’ species strains sharing > 98.65% sequence identity of their full or nearly full 16S rRNA gene sequences, obtained with at least 2-fold coverage of the sequence, compared with those of the reference strain of such species [4]. The officially published ‘Candidatus Phytoplasma’ species are 49; however, they do not cover all the relevant biodiversity, especially in reference to differential geographic distribution and/or host species. Therefore, the differentiation in ribosomal groups and subgroups [12] is still valuable and should be used to be able to work on their epidemiology and prevention in the different areas of the world. The main distribution of strains is tightly related to the geographic areas and to the dissemination performed by propagation materials, such as cuttings and seeds, that are also infected, even if only in low percentages (1–3%).
4. Relationship between Phytoplasma Symptomatology and Classification
Together with the study on the diseases associated with the presence of these bacteria, the first step was to give them a name according to the diverse disease in which the association was detected with specific phytoplasmas. Today, 30 years after this exercise started it appears clear that the molecular diversity described in phytoplasmas (49 ‘Candidatus Phytoplasma’ species and about 200 ribosomal subgroups) using the 16S ribosomal gene as basic standard is only in some cases related to a differential symptomatology. Identical symptoms are associated with different phytoplasmas and vice versa. Moreover, phytoplasmas associated with decline symptoms in some species could be associated with phyllody/virescence in others, such as ‘Ca. P. solani’ infecting potatoes, tomatoes, and grapevine. Therefore, contrary to the other plant pathogens it is necessary to verify the pathogen identity by molecular tools on a case-by-case basis; however, at the same time, it is of utmost importance to also recognize the symptoms associated with the presence of the phytoplasmas in order to appropriately manage the disease. This review of the main symptoms and several associated phytoplasmas worldwide is aimed at helping the recognition of the presence of these bacteria in plants, further clarifying their relationship with the host plants. This feature is, however, not stable over time also in the same plant species, considering the never-ending mechanism of patho-adaption that is part of life also in microorganisms; pathogens are special microorganisms that are simply looking for new ecological niches to ensure their survival and do not aim to destroy or kill the hosts.
4.1. Shoot Proliferation and Witches' Broom
Diseases with symptoms of witches’ broom can be caused by basidiomycetes but could also be associated with the presence of phytoplasmas. In both cases they are economically important in a number of crop plant species, including the cocoa tree, jujube, citrus, and apple and timber trees, such as poplar, Melia azedarach, and paulownia (Figure 3). Among woody species, this malformation is almost always associated with the presence of specific phytoplasmas, such as in apple (‘Ca. P. mali), lime (‘Ca. P. aurantifolia’), lilac (‘Ca. P. fraxini’), paulownia (‘Ca. P. asteris’), almond (‘Ca. P. phoenicium’), Juniperus (16SrIX-E), walnut (16SrIII-G), Balanites triflora (‘Ca. P. balanites’), spartium (‘Ca. P. spartii’), black alder (16SrX-E), hibiscus (‘Ca. P. brasiliense’), Guazuma spp. (16SrXV-B), chestnut (‘Ca. P. castaneae’), Cassia italica (‘Ca. P. omanense’), and salt cedar (‘Ca. P. tamaricis’). In herbaceous host plants, the presence of witches’ broom was reported in diverse species, some of them as hosts of new phytoplasma strains (Table 1), such as strawberry, peanut, cactus, tabebuja, tomatillo, chayote, black raspberry, erigeron, alfalfa, and pigeon pea.
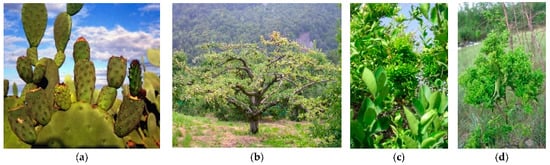
Figure 3. Cactus pear (Opuntia ficus-indica) proliferation (a), apple proliferation (b), citrus witches’ broom (c), and jujube witches’ broom (d) are associated with the presence of phytoplasmas in diverse areas of the world.
Table 1. Molecular diversity and geographic distribution of selected phytoplasmas belonging to different ribosomal groups/‘Candidatus Phytoplasma’ species (marked by different color) associated with witches’ broom symptoms.
| Disease (Acronym) | Continent | 16Sr Subgroups | ‘Candidatus Phytoplasma’ Species |
GenBank Accession Number | References |
|---|---|---|---|---|---|
| Aster yellows w. b. (AY-WB) | America | 16SrI-A | ‘Ca. P. asteris’ | NC_007716 | [13] |
| Paulownia w. b. (PaWB) | Asia | 16SrI-D | AY265206 | [14] | |
| Strawberry witches’ broom (STRAWB1), (STRAWB2) | America | 16SrI-I / -K | U96614, U96616 | [15] | |
| Peach rosette-like (PRU0382) | America | 16SrI-W | HQ450211 | [16] | |
| Peanut witches’ broom (PnWB) | America | 16SrII-A | L33765 | [17] | |
| Lime witches’ broom (WBDL) | Asia | 16SrII-B | ‘Ca. P. aurantifolia’ | U15442 | [18] |
| Cactus witches’ broom (CWB) | Asia | 16SrII-G to -L | EU099568, EU099552, EU099569, EU099572, EU099551, EU099546, EF647744 | [19] | |
| Tabebuia witches’ broom | America | 16SrII-O | [20] | ||
| Tomatillo witches’ broom | America | 16SrII-T | U125185 | [21] | |
| Walnut witches’ broom (WWB) | America | 16SrIII-G | AF190226, AF190227 | [22] | |
| Poinsettia branch-inducing (PoiBI) | Europe, America |
16SrIII-H | AF190223 | [22] | |
| Chayote w. b. (ChWBIII) | America | 16SrIII-J | AF147706 | [23] | |
| Black raspberry w. b. (BRWB7) | America | 16SrIII-Q | AF302841 | [24] | |
| Conyza witches’ broom | America | 16SrIII-X | KC412026 | [25] | |
| Jujube witches’ broom (JWB-G1) | Asia | 16SrV-B | ‘Ca. P. ziziphi’ | AB052876 | [26] |
| Balanites triflora w. b. (BltWB) | Asia | 16SrV-F | Ca. P. balanitae’ | AB689678 | [27] |
| Korean jujube witches’ broom | Asia | 16SrV-G | AB052879 | [26] | |
| Bischofia polycarpa witches’ broom | Asia | 16SrV-H | KJ452547 | [28] | |
| Blackberry witches’ broom | Europe | 16SrV-I | KR233473 | [29] | |
| Clover proliferation (CP) | America | 16SrVI-A | ‘Ca. P. trifolii’ | AY390261 | [30] |
| Erigeron witches’ broom (ErWB) | America | 16SrVII-B | AY034608 | [31] | |
| Argentinian alfalfa w.b. (ArAWB) | America | 16SrVII-C | AY147038 | [32] | |
| Erigeron w. b. (EboWB-Br0) | America | 16SrVII-D | KJ831066 | [33] | |
| Loofah witches’ broom (LufWB) | Asia | 16Sr VIII-A | ‘Ca. P. luffae’ | AF086621 | [34] |
| Pigeon pea w. b. (PPWB) | America | 16SrIX-A | AF248957 | [35] | |
| Almond witches’ broom (AlWB) | Asia | 16SrIX-B/-D | ‘Ca. P. phoenicium’ | AF515636, AF515637 | [36] |
| Juniperus witches’ broom | America | 16SrIX-E | GQ925918 | [37] | |
| Almond and stone fruit witches’ broom (N27-2), (A1-1) | Asia | 16SrIX-F/-G | ‘Ca. P. phoenicium’ | HQ407532, HQ407514 | [38] |
| Apple proliferation (AP) | Europe, Asia | 16SrX-A | ‘Ca. P. mali’ | AJ542541 | [39] |
| Spartium witches’ broom (SpaWB) | Europe | 16SrX-D | ‘Ca. P. spartii’ | X92869 | [40] |
| Black alder w. b. (BAWB, BWB) | Europe | 16SrX-E | X76431 | [41] | |
| Hibiscus witches’ broom (HibWB) | America, Asia | 16SrXV-A | ‘Ca. P. brasiliense’ | AF147708 | [42] |
| Guazuma w. b. (GWB) | America | 16SrXV-B | HQ258882 | [43] | |
| Chestnut witches’ broom | Asia | 16SrXIX-A | ‘Ca. P. castaneae’ | AB054986 | [44] |
| Rhamnus witches’ broom | Europe | 16SrXX-A | ‘Ca. P. rhamni’ | AJ583009 | [40] |
| Weeping tea witches’ broom | Oceania | 16SrXXV-A * | AF521672 | [45] | |
| Cassia w. b. (CaWB) | Asia | 16SrXXIX-A | ‘Ca. P. omanense’ | EF666051 | [46] |
| Bindweed witches’ broom (RBiWB) | Asia | 16SrXXIX-B | KY047493 | [47] | |
| Salt cedar witches’ broom | Asia | 16SrXXX-A | ‘Ca. P. tamaricis’ | FJ432664 | [48] |
w. b., witches’ broom; *, described as sequence deposited in GenBank only.
The excessive shoot proliferation results in poor or no fruit production and severely reduces the cultivation of some of these crops. Citrus in the Arabian Peninsula, jujube in China, and apple proliferation in Europe are some of the most severe cases that greatly reduce the possibility to produce and commercialize popular fruits. This modification is due to the loss of apical dominance of the shoots linked to disorders in the hormone balance.
4.2. Stunting and Little Leaf
Stunting in plants could be due to virus or phytoplasma presence; however, it must also be verified that glyphosate or similar pesticides were not applied in the area in which these malformations are present in plants in the past years, since this can produce indistinguishable symptoms (Figure 4). The presence of phytoplasmas is reported in several plant species enclosing small fruits, vegetables, corn, and soybean; in some cases, these bacteria were associated with the presence of little leaf or stunting also in trees, such as cherry, eucalyptus, and Sophora japonica [49] (Table 2). In strawberries the case Fragaria multicipita was discovered to be not a true species, but just a cloned phytoplasma-infected genotype [15]. The hormone imbalance, according with the diverse infected species, is usually present and the transportation of starch and other metabolites for the appropriate development is very often impaired.

Figure 4. Sophora japonica stunting and yellows (a); Eucalyptus little leaf (b), periwinkle little leaf (c), and corn stunting (d).
Table 2. Molecular diversity and geographic distribution of selected phytoplasmas belonging to different ribosomal groups/‘Candidatus Phytoplasma’ species (marked by different color) associated with little leaf and stunting symptoms.
| Disease (Acronym) | Continent | 16Sr Subgroups |
‘Candidatus Phytoplasma’ Species |
GenBank Accession Number | References |
|---|---|---|---|---|---|
| Blue dwarf wheat (BDW) | Asia | 16SrI-C | ‘Ca. P. tritici’ | DQ078304 | [50] |
| Blueberry stunt (BBS3) | America | 16SrI-E | AY265213 | [14] | |
| Cherry little leaf (ChLL) | Europe | 16SrI-Q | AY034089 | [51] | |
| Pepper little leaf (PeLL) | America | 16SrI-S | DQ092321 | [52] | |
| Tomato little leaf (ToLL) | America | 16SrI-T | DQ375238 | [52] | |
| Vasconcellea cundinamarcensis little leaf | China | 16SrII-U | KP057205 | [53] | |
| Spiraea stunt (SP1) | America | 16SrIII-E | AF190228 | [23] | |
| Heterothalamus little leaf (HetLL) | America | 16SrIII-W | KC412029 | [26] | |
| Broccoli stunt (BSP-21) | America | 16SrIII-Z | JX626327 | [22] | |
| Rubus stunt (RuS) | Europe | 16SrV-E | ‘Ca. P. rubi’ | AY197648 | [54] |
| Fragaria multicipita, multiplier disease | America | 16SrVI-B | AF190224 | [15] | |
| Periwinkle little leaf (PLL-Bd) | Asia | 16SrVI-D | AF228053 | [55] | |
| Portulaca little leaf (PLL-Ind) | Asia | 16SrVI-H | EF651786 | [56] | |
| Soybean stunt (SoyST1c1) | America | 16SrXXXI-A | ‘Ca. P. costaricanum’ | HQ225630 | [57] |
4.3. Phyllody and Virescence
The transformation of different plant organs into leaves is a very relevant symptom among those associated with phytoplasma presence and is known as phyllody; this type of malformation could also be due to the application of pesticides based on hormone-like molecules. The virescence is the change of the color of flowers to green, which is due to phytoplasma presence, but in some cases the diagnostics can be tricked by the existence of flowers that are green and the presence of genetic factors modifying the anthocian distribution in the plant, as can be seen in a Chinese variety of rose and in some special clones of periwinkle (Figure 5). The most relevant phytoplasma-associated diseases are reported in flowering species for commercialization; however, virescence is also present in horticultural and seed crops, such as tomatoes, cabbages, strawberries, and clover, among several other species (Table 3).

Figure 5. Virescence in gladiolus (a) and in periwinkle (b); phyllody in echinaea (c), rose (d), and strawberry (e). The rose flowers are showing virescence and phyllody due to genetics, rather than the phytoplasma presence in all the others.
Table 3. Molecular diversity and geographic distribution of selected phytoplasmas belonging to different ribosomal groups/‘Candidatus Phytoplasma’ species (marked by different color) and associated with phyllody and virescence symptoms.
| Disease (Acronym) | Continent | 16Sr Subgroups |
‘Candidatus Phytoplasma’ Species |
GenBank Accession Number | References |
|---|---|---|---|---|---|
| Clover phyllody (CPh) | America | 16SrI-C | AF222065 | [15] | |
| Faba bean phyllody (FBP) | Asia, Africa | 16SrII-C | X83432 | [58] | |
| Pichris echioides phyllody (PEY) | Europe | 16SrII-E | Y16393 | [58] | |
| Cotton phyllody (CoP) | Africa | 16SrII-F | EF186827 | [59] | |
| Strawberry leafy fruit (SLF) | America | 16SrIII-K | AF274876 | [15] | |
| Dandelion virescence (DanVir) | Europe | 16SrIII-O/-P | AF370120, AF370119 | [60] | |
| Heterothalamus little leaf (HetLL) | America | 16SrIII-W | KC412029 | [26] | |
| Centarurea solstitialis virescence (CSVI) | Europe | 16SrVI-E | AY270156 | [61] | |
| Catharanthus phyllody (CPS) | Africa | 16SrVI-F | EF186819 | [59] | |
| Naxos periwinkle virescence (NAXOS) | Europe, Asia, America | 16SrIX-C | HQ589191 | [62] | |
| Sarsoon phyllody | Asia | 16SrIX-H | KU892213 | [63] | |
| Japanese hydrangea phyllody | Asia | 16SrXII-D | ‘Ca. P. japonicum’ | AB010425 | [64] |
| Mexican periwinkle virescence (MPV) | America | 16SrXIII-A | ‘Ca. P. hispanicum’ | AF248960 | [65] |
| Strawberry green petal (STRAWB2) | America | 16SrXIII-B | U96616 | [15] | |
| Malaysian periwinkle virescence (MaPV) | Asia | 16SrXXXII-A | ‘Ca. P. malaysianum’ | EU371934 | [66] |
This entry is adapted from the peer-reviewed paper 10.3390/plants11111425
References
- McCann, H.C.; Li, L.; Liu, Y.; Li, D.; Pan, H.; Zhong, C.; Rikkerink, E.; Templeton, M.D.; Straub, C.; Colombi, E.; et al. Origin and evolution of the kiwifruit canker pandemic. Gen. Biol. Evol. 2017, 9, 932–944.
- Bové, J.-M. Huanglongbing: A destructive, newly-emerging, century-old disease of citrus. J. Plant Pathol. 2006, 88, 7–37.
- IRPCM. ‘Candidatus Phytoplasma’, a taxon for the wall-less, non-helical prokaryotes that colonise plant phloem and insects. Int. J. Syst. Evol. Microbiol. 2004, 54, 1243–1255.
- Bertaccini, A.; Arocha-Rosete, Y.; Contaldo, N.; Duduk, B.; Fiore, N.; Montano, H.G.; Kube, M.; Kuo, C.-H.; Martini, M.; Oshima, K.; et al. Revision of the ‘Candidatus Phytoplasma’ species description guidelines. Int. J. Syst. Evol. Microbiol. 2022, 74, 005353.
- Namba, S. Molecular and biological properties of phytoplasmas. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2019, 95, 401–418.
- Doi, Y.; Teranaka, M.; Yora, K.; Asuyama, H. Mycoplasma or PLT grouplike microrganisms found in the phloem elements of plants infected with mulberry dwarf, potato witches’ broom, aster yellows or pawlownia witches’ broom. Ann. Phytopath. Soc. Jpn. 1967, 33, 259–266.
- Contaldo, N.; Bertaccini, A.; Paltrinieri, S.; Windsor, H.M.; Windsor, G.D. Axenic culture of plant pathogenic phytoplasmas. Phytopath. Medit. 2012, 51, 607–617.
- Contaldo, N.; Satta, E.; Zambon, Y.; Paltrinieri, S.; Bertaccini, A. Development and evaluation of different complex media for phytoplasma isolation and growth. J. Microbiol. Meth. 2016, 127, 105–110.
- Contaldo, N.; D’Amico, G.; Paltrinieri, S.; Diallo, H.A.; Bertaccini, A.; Arocha Rosete, Y. Molecular and biological characterization of phytoplasmas from coconut palms affected by the lethal yellowing disease in Africa. Microbiol. Res. 2019, 223-225, 51–57.
- Luis Pantoja, M.; Paredes-Tomás, C.; Uneau, Y.; Myrie, W.; Morillon, R.; Satta, E.; Contaldo, N.; Pacini, F.; Bertaccini, A. Identification of ‘Candidatus Phytoplasma’ species in “huanglongbing” infected citrus orchards in the Caribbean. Eur. J. Plant Pathol. 2021, 160, 185–198.
- Betancourt, C.; Pardo, J.; Muñoz, J.; Alvarez, E. Isolation of phytoplasmas associated to frogskin disease in cassava. Rev. UDCA Actual. Divulg. Cient. 2019, 22, e1177.
- Lee, I.-M.; Gundersen-Rindal, D.E.; Davis, R.E.; Bartoszyk, I.M. Revised classification scheme of phytoplasmas based on RFLP analyses of 16S rRNA and ribosomal protein gene sequences. Int. J. Syst. Evol. Microbiol. 1998, 48, 1153–1169.
- Bai, X.; Zhang, J.; Ewing, A.; Miller, S.A.; Radek, A.J.; Shevchenko, D.V.; Tsukerman, K.; Walunas, T.; Lapidus, A.; Campbell, J.W.; et al. Living with genome instability: The adaptation of phytoplasmas to diverse environments of their insect and plant hosts. J. Bacteriol. 2006, 188, 3682–3696.
- Lee, I.-M.; Gundersen-Rindal, D.E.; Davis, R.E.; Bottner, K.D.; Marcone, C.; Seeműller, E. ‘Candidatus Phytoplasma asteris’, a novel taxon associated with aster yellows and related diseases. Int. J. Syst. Evol. Microbiol. 2004, 54, 1037–1048.
- Jomantiene, R.; Davis, R.E.; Maas, J.; Dally, E.L. Classification of new phytoplasmas associated with diseases of strawberry in Florida, based on analysis of 16S rRNA and ribosomal protein gene operon sequences. Int. J. Syst. Evol. Microbiol. 1998, 48, 269–277.
- Arocha-Rosete, Y.; Zunnoon-Khan, S.; Krukovets, I.; Crosby, W.; Scott, J.; Bertaccini, A.; Michelutti, R. Identification and molecular characterization of the phytoplasma associated with peach rosette-like disease at the Canadian clonal Genebank based on the 16S rRNA gene analysis. Can. J. Plant Pathol. 2011, 33, 127–134.
- Gundersen, D.E.; Lee, I.-M.; Rehner, S.A.; Davis, R.E.; Kingsbury, D.T. Phylogeny of mycoplasmalike organisms (phytoplasmas): A basis for their classification. J. Bacteriol. 1994, 176, 5244–5254.
- Zreik, L.; Carle, P.; Bové, J.-M.; Garnier, M. Characterization of the mycoplasmalike organism associated with witches’ broom disease of lime and proposition of a ‘Candidatus’ taxon for the organism, ‘Candidatus Phytoplasma aurantifolia’. Int. J. Syst. Bacteriol. 1995, 45, 449–453.
- Cai, H.; Wei, W.; Davis, R.E.; Chen, H.; Zhao, Y. Genetic diversity among phytoplasmas infecting Opuntia species: Virtual RFLP analysis identifies new subgroups in the peanut witches’ broom phytoplasma group. Int. J. Syst. Evol. Microbiol. 2008, 58, 1448–1457.
- Mafia, R.G.; Barreto, R.W.; Vanetti, C.A.; Hodgetts, J.; Dickinson, M.; Alfenas, A.C. A phytoplasma is associated with witches’ broom disease of Tabebuia pentaphylla in Brazil. New Dis. Rep. 2007, 15, 49.
- Perez-López, E.; Luna-Rodríguez, M.; Olivier, C.Y.; Dumonceaux, T.J. The underestimated diversity of phytoplasmas in Latin America. Int. J. Syst. Evol. Microbiol. 2016, 66, 492–513.
- Davis, R.E.; Zhao, Y.; Dally, E.L.; Lee, I.-M.; Jomantiene, R.; Douglas, S. ‘Candidatus Phytoplasma pruni’, a novel taxon associated with X-disease of stone fruits, Prunus spp.: Multilocus characterization based on 16S rRNA, secY, and ribosomal protein genes. Int. J. Syst. Evol. Microbiol. 2013, 63, 766–776.
- Montano, H.G.; Davis, R.E.; Dally, E.L.; Pimentel, J.P.; Brioso, P.S.T. Identification and phylogenetic analysis of a new phytoplasma from diseased chayote in Brazil. Plant Dis. 2000, 84, 429–436.
- Davis, R.E.; Dally, E.L.; Converse, R.H. Molecular identification of a phytoplasma associated with witches’-broom disease of black raspberry in Oregon and its classification in group 16SrIII, new subgroup Q. Plant Dis. 2001, 85, 1121.
- Galdeano, E.; Guzmán, F.A.; Fernández, F.; Conci, R.G. Genetic diversity of 16SrIII group phytoplasmas in Argentina. Predominance of subgroups 16SrIII-J and B and two new subgroups 16SrIII-W and X. Eur. J. Plant Pathol. 2013, 137, 753–764.
- Jung, H.-Y.; Sawayanagi, T.; Kakizawa, S.; Nishigawa, H.; Wei, W.; Oshima, K.; Miyata, S.; Ugaki, M.; Hibi, T.; Namba, S. ‘Candidatus Phytoplasma ziziphi’, a novel phytoplasma taxon associated with jujube witches’ broom disease. Int. J. Syst. Evol. Microbiol. 2003, 53, 1037–1041.
- Win, N.K.K.; Lee, S.-Y.; Bertaccini, A.; Namba, S.; Jung, H.-Y. ‘Candidatus Phytoplasma balanitae’ associated with witches’ broom disease of Balanites triflora. Int. J. Syst. Evol. Microbiol. 2013, 63, 636–640.
- Lai, F.; Song, C.S.; Ren, Z.G.; Lin, C.L.; Xu, Q.C.; Li, Y.; Piao, C.G.; Yu, S.S.; Guo, M.W.; Tian, G.Z. Molecular characterization of a new member of the 16SrV group of phytoplasma associated with Bischofia polycarpa (Levl.) Airy Shaw witches’ broom disease in China by a multiple gene-based analysis. Austral. Plant Pathol. 2014, 43, 557–569.
- Fránová, J.; de Sousa, E.; Mimoso, C.; Cardoso, F.; Contaldo, N.; Paltrinieri, S.; Bertaccini, A. Multigene characterization of a new ‘Candidatus Phytoplasma rubi’-related strain associated with blackberry witches’ broom in Portugal. Int. J. Syst. Evol. Microbiol. 2016, 66, 1438–1446.
- Hiruki, C.; Wang, K. Clover proliferation phytoplasma: ‘Candidatus Phytoplasma trifolii’. Int. J. Syst. Evol. Microbiol. 2004, 54, 1349–1353.
- Barros, T.S.L.; Davis, R.E.; Resende, R.O.; Dally, E.L. Erigeron witches’ broom phytoplasma in Brazil represents new subgroup VII-B in 16S rRNA gene group VII, the ash yellows phytoplasma group. Plant Dis. 2002, 86, 1142–1148.
- Conci, L.; Meneguzzi, N.; Galdeano, E.; Torres, L.; Nome, C.; Nome, S. Detection and molecular characterisation of an alfalfa phytoplasma in Argentina that represents a new subgroup in the 16S rDNA ash yellows group (‘Candidatus Phytoplasma fraxini’). Eur. J. Plant Pathol. 2005, 113, 255–265.
- Flôres, D.; Amaral Mello, A.O.; Pereira, T.B.C.; Rezende, J.A.M.; Bedendo, I.P. A novel subgroup 16SrVII-D phytoplasma identified in association with erigeron witches’ broom. Int. J. Syst. Evol. Microbiol. 2015, 65, 2761–2765.
- Davis, R.E.; Zhao, Y.; Wei, W.; Dally, E.L.; Lee, I.-M. ‘Candidatus Phytoplasma luffae’, a novel taxon associated with witches’ broom disease of loofah, Luffa aegyptica Mill. Int. J. Syst. Evol. Microbiol. 2017, 67, 3127–3133.
- Gundersen, D.E.; Lee, I.-M.; Schaff, D.A.; Harrison, N.A.; Chang, C.J.; Davis, R.E.; Kinsbury, D.T. Genomic diversity among phytoplasma strains in 16S rRNA group I (aster yellows and related phytoplasmas) and III (X-disease and related phytoplasmas). Int. J. Syst. Bacteriol. 1996, 46, 64–75.
- Verdin, E.; Salar, P.; Danet, J.-L.; Choueiri, E.; Jreijiri, F.; El Zammar, S.; Gélie, B.; Bové, J.-M.; Garnier, M. ‘Candidatus Phytoplasma phoenicium’ sp. nov., a novel phytoplasma associated with an emerging lethal disease of almond trees in Lebanon and Iran. Int. J. Syst. Evol. Microbiol. 2003, 53, 833–838.
- Davis, R.E.; Dally, E.; Zhao, Y.; Lee, I.-M.; Jomantiene, R.; Detweiler, A.J.; Putnam, M.L. First report of a new subgroup 16SrIX-E (‘Candidatus Phytoplasma phoenicium’-related) phytoplasma associated with juniper witches’ broom disease in Oregon, USA. Plant Pathol. 2010, 59, 1161.
- Molino Lova, M.; Quaglino, F.; Abou-Jawdah, Y.; Choueiri, E.; Sobh, H.; Casati, P.; Tedeschi, R.; Alma, A.; Bianco, P.A. Identification of new 16SrIX subgroups, -F and -G, among ‘Candidatus Phytoplasma phoenicium’ strains infecting almond, peach and nectarine in Lebanon. Phytopath. Medit. 2011, 50, 273–282.
- Seemüller, E.; Schneider, B. ‘Candidatus Phytoplasma mali’, ‘Candidatus Phytoplasma pyri’ and ‘Candidatus Phytoplasma prunorum’, the causal agents of apple proliferation, pear decline and European stone fruit yellows, respectively. Int. J. Syst. Evol. Microbiol. 2004, 54, 1217–1226.
- Marcone, C.; Gibb, K.S.; Streten, C.; Schneider, B. ‘Candidatus Phytoplasma spartii’, ‘Candidatus Phytoplasma rhamni’ and ‘Candidatus Phytoplasma allocasuarinae’, respectively associated with Spartium witches’ broom, buckthorn witches’ broom and Allocasuarina yellows diseases. Int. J. Syst. Evol. Microbiol. 2004, 54, 1025–1029.
- Seemüller, E.; Schneider, B.; Maurer, R.; Ahrens, U.; Daire, X.; Kison, H.; Lorenz, K.; Firrao, G.; Avinent, L.; Sears, B.B.; et al. Phylogenetic classification of phytopathogenic mollicutes by sequence analysis of 16S ribosomal DNA. Int. J. Syst. Bacteriol. 1994, 44, 440–446.
- Montano, H.G.; Davis, R.E.; Dally, E.L.; Hogenhout, S.; Pimentel, J.P.; Brioso, P.S. ‘Candidatus Phytoplasma brasiliense’, a new phytoplasma taxon associated with hibiscus witches’ broom disease. Int. J. Syst. Evol. Microbiol. 2001, 51, 1109–1118.
- Villalobos, W.; Martini, M.; Garita, L.; Muñoz, M.; Osler, R.; Moreira, L. Guazuma ulmifolia (Sterculiaceae), a new natural host of 16SrXV phytoplasma in Costa Rica. Trop. Plant Pathol. 2011, 36, 2.
- Jung, H.-Y.; Sawayanagi, T.; Kakizawa, S.; Nishigawa, H.; Miyata, S.; Oshima, K.; Ugaki, M.; Lee, J.T.; Hibi, T.; Namba, S. ‘Candidatus Phytoplasma castaneae’, a novel phytoplasma taxon associated with chestnut witches’ broom disease. Int. J. Syst. Evol. Microbiol. 2002, 52, 1543–1549.
- Wei, W.; Davis, R.E.; Lee, I.-M.; Zhao, Y. Computer-simulated RFLP analysis of 16S rRNA genes: Identification of ten new phytoplasma groups. Int. J. Syst. Evol. Microbiol. 2007, 57, 1855–1867.
- Al-Saady, N.A.; Khan, A.J.; Calari, A.; Al-Subhi, A.M.; Bertaccini, A. ‘Candidatus Phytoplasma omanense’, associated with witches’ broom of Cassia italica (Mill.) Spreng in Oman. Int. J. Syst. Evol. Microbiol. 2008, 58, 461–466.
- Esmailzadeh Hosseini, S.A.; Salehi, M.; Mirchenari, S.M.; Contaldo, N.; Paltrinieri, S.; Bertaccini, A. Occurrence of a ‘Candidatus Phytoplasma omanense’-related strain in bindweed showing a witches’ broom disease in Iran. Phytopath. Moll. 2016, 6, 87–92.
- Zhao, Y.; Sun, Q.; Wei, W.; Davis, R.E.; Wu, W.; Liu, Q. ‘Candidatus Phytoplasma tamaricis’, a novel taxon discovered in witches’-broom-diseased salt cedar (Tamarix chinensis Lour.). Int. J. Syst. Evol. Microbiol. 2009, 59, 2496–2504.
- Duduk, B.; Tian, J.B.; Contaldo, N.; Fan, X.P.; Paltrinieri, S.; Chen, Q.F.; Zhao, Q.F.; Bertaccini, A. Occurrence of phytoplasmas related to “stolbur” and to ‘Candidatus Phytoplasma japonicum’ in woody host plants in China. J. Phytopath. 2010, 158, 100–104.
- Zhao, Y.; Wei, W.; Davis, R.E.; Lee, I.-M.; Bottner-Parker, K.D. The agent associated with blue dwarf disease in wheat represents a new phytoplasma taxon, ‘Candidatus Phytoplasma tritici’. Int. J. Syst. Evol. Microbiol. 2021, 71, 004604.
- Valiunas, D.; Jomantiene, R.; Davis, R.E. A ‘Candidatus Phytoplasma asteris’-related phytoplasma associated with cherry little leaf disease represents a new subgroup, 16SrI-Q. Phytopathology 2005, 95, S106.
- Santos-Cervantes, M.E.; Chávez-Medina, J.A.; Acosta-Pardini, J.; Flores-Zamora, G.L.; Méndez-Lozano, J.; Leyva-López, N.E. Genetic diversity and geographical distribution of phytoplasmas associated with potato purple top disease in Mexico. Plant Dis. 2010, 94, 388–395.
- Yang, Y.; Jiang, L.; Che, H.; Cao, X.; Luo, D. Identification of a novel subgroup 16SrII-U phytoplasma associated with papaya little leaf disease. Int. J. Syst. Evol. Microbiol. 2016, 66, 3485–3491.
- Malembic-Maher, S.; Salar, P.; Filippin, L.; Carle, P.; Angelini, E.; Foissac, X. Genetic diversity of European phytoplasmas of the 16SrV taxonomic group and proposal of ‘Candidatus Phytoplasma rubi’. Int. J. Syst. Evol. Microbiol. 2011, 61, 2129–2134.
- Siddique, A.B.M.; Agrawal, G.K.; Alam, N.; Krishina Reddy, M. Electron microscopy and molecular characterization of phytoplasmas associated with little leaf disease of brinjal (Solanum melongena) and periwinkle (Catharanthus roseus) in Bangladesh. J. Phytopath. 2001, 149, 237–244.
- Samad, A.; Ajayakumar, P.V.; Shasany, A.K.; Gupta, M.K.; Alam, M.; Rastogi, S. Occurrence of a clover proliferation (16SrVI) group phytoplasma associated with little leaf disease of Portulaca grandiflora in India. Plant Dis. 2008, 92, 832.
- Lee, I.-M.; Bottner-Parker, K.D.; Zhao, Y.; Villalobos, W.; Moreira, L. ‘Candidatus Phytoplasma costaricanum’ a novel phytoplasma associated with an emerging disease in soybean (Glycine max). Int. J. Syst. Evol. Microbiol. 2011, 61, 2822–2826.
- Seemüller, E.; Marcone, C.; Lauer, U.; Ragozzino, A.; Göschl, M. Current status of molecular classification of the phytoplasmas. J. Plant Pathol. 1998, 80, 3–26.
- Martini, M.; Lee, I.-M.; Bottner, K.D.; Zhao, Y.; Botti, S.; Bertaccini, A.; Harrison, N.A.; Carraro, L.; Marcone, C.; Khan, J.; et al. Ribosomal protein gene-based phylogeny for finer differentiation and classification of phytoplasmas. Int. J. Syst. Evol. Microbiol. 2007, 57, 2037–2051.
- Jomantiene, R.; Maas, J.L.; Takeda, F.; Davis, R.E. Molecular identification and classification of strawberry phylloid fruit phytoplasma in group 16SrI, new subgroup. Plant Dis. 2002, 86, 920.
- Faggioli, F.; Pasquini, G.; Lumia, V.; Campobasso, G.; Widmer, T.L.; Quimby, P.C. Molecular identification of a new member of the clover proliferation phytoplasma group (16SrVI) associated with yellow starthistle virescence in Italy. Eur. J. Plant Pathol. 2004, 110, 353–360.
- Duduk, B.; Mejia, J.F.; Calari, A.; Bertaccini, A. Identification of 16SrIX group phytoplasmas infecting Colombian periwinkles and molecular characterization on several genes. In Proceedings of the 17th IOM Congress, Tienjin, China, 6–11 July 2008; p. 83.
- Ahmad, J.N.; Ahmad, S.J.N.; Irfan, M.; Paltrinieri, S.; Contaldo, N.; Bertaccini, A. Molecular detection, identification, characterization and transmission study of sarsoon phyllody in Punjab–Pakistan associated with phytoplasmas affiliated to the new subgroup 16SrIX-H. Eur. J. Plant Pathol. 2017, 149, 117–125.
- Sawayanagi, T.; Horikoshi, N.; Kanehira, T.; Shinohara, M.; Bertaccini, A.; Cousin, M.-T.; Hiruki, C.; Namba, S. ‘Candidatus Phytoplasma japonicum’, a new phytoplasma taxon associated with Japanese hydrangea phyllody. Int. J. Syst. Evol. Microbiol. 1999, 49, 1275–1285.
- Davis, R.E.; Harrison, N.A.; Zhao, Y.; Wei, W.; Dally, E.L. ‘Candidatus Phytoplasma hispanicum’, a novel taxon associated with Mexican periwinkle virescence disease of Catharanthus roseus. Int. J. Syst. Evol. Microbiol. 2016, 66, 3463–3467.
- Nejat, N.; Vadamalai, G.; Davis, R.E.; Harrison, N.A.; Sijam, K.; Dickinson, M.; Abdullah, S.N.A.; Zhao, Y. ‘Candidatus Phytoplasma malaysianum’, a novel taxon associated with virescence and phyllody of Madagascar periwinkle (Catharanthus roseus). Int. J. Syst. Evol. Microbiol. 2013, 63, 540–548.
This entry is offline, you can click here to edit this entry!
