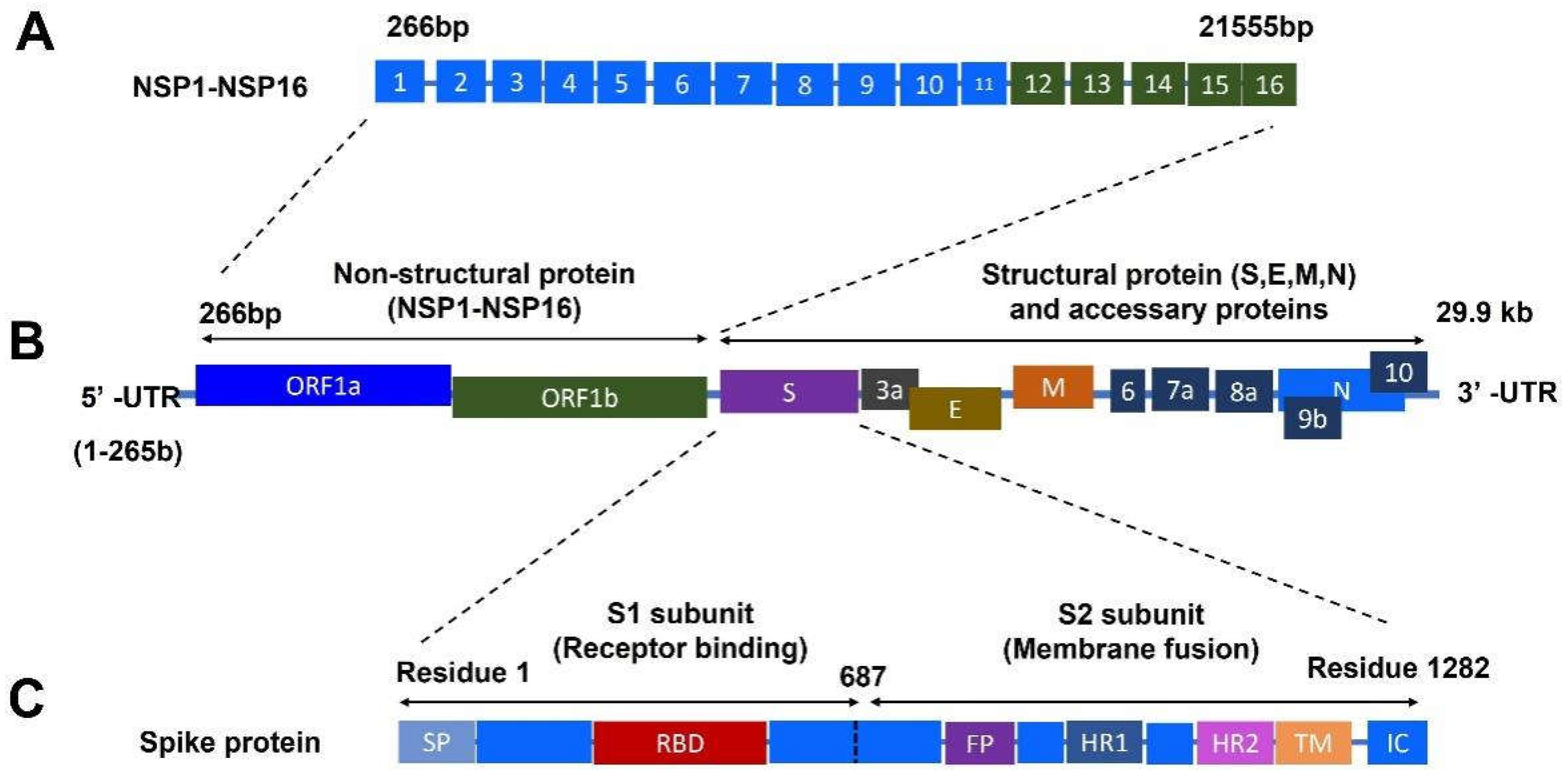
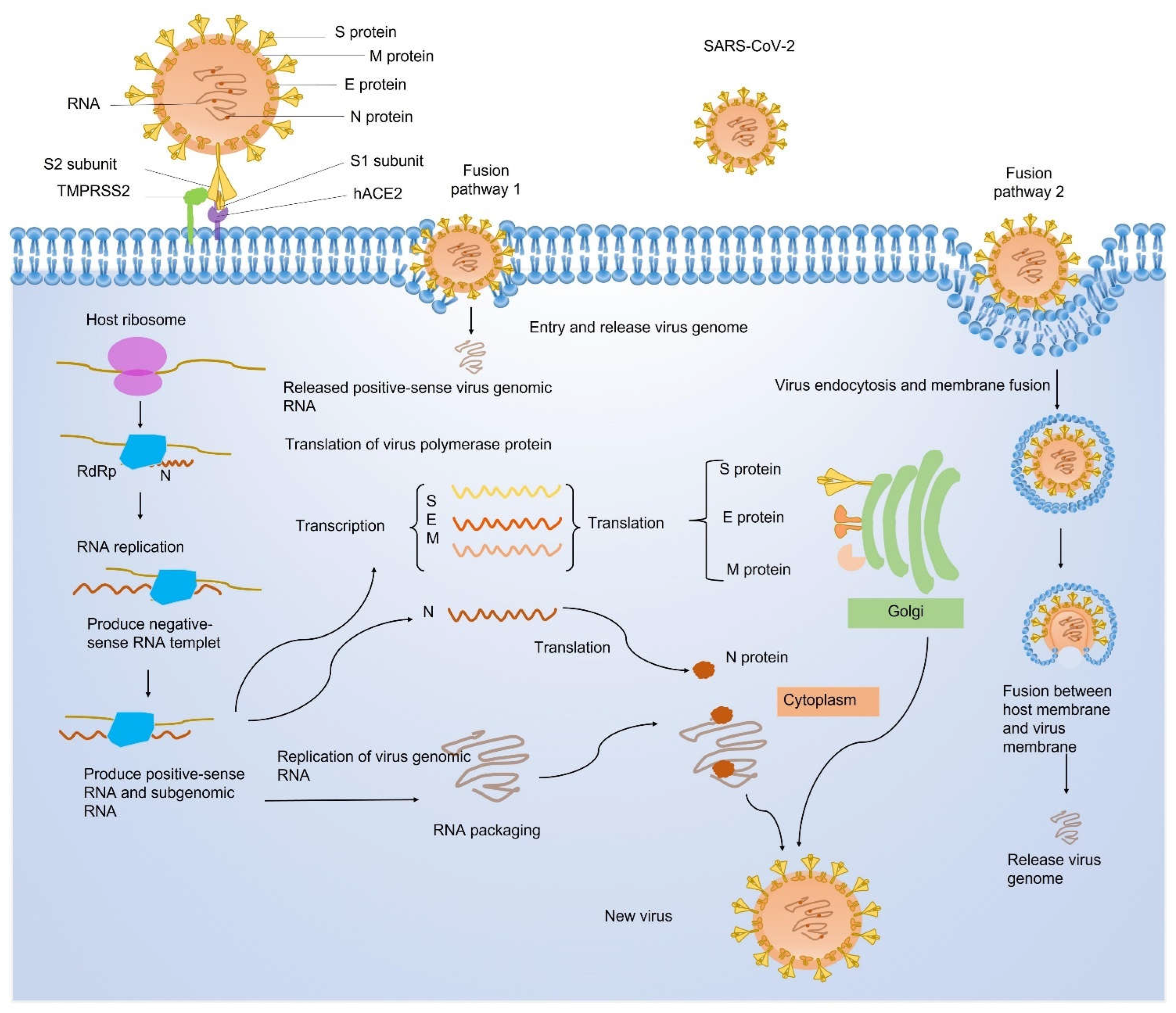
Coronavirus disease 2019 (COVID-19) caused by the infection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has become the most severe health crisis, causing extraordinary economic disruption worldwide. SARS-CoV-2 is a single-stranded RNA-enveloped virus. The process of viral replication and particle packaging is finished in host cells. Viral proteins, including both structural and nonstructural proteins, play important roles in the viral life cycle, which also provides the targets of treatment. Therefore, a better understanding of the structural function of virus proteins is crucial to speed up the development of vaccines and therapeutic strategies. The structure-function correlation of viral proteins provides a fundamental rationale for vaccine development and targeted therapy.
- COVID-19
- SARS-CoV-2
- nonstructural proteins
- structural proteins
- vaccines
- therapy
1. Introduction of COVID-19
1.1. COVID-19 Pandemic
2. Vaccine Approach to Prevent Infection
2.1. SARS-CoV-2 Inactivated Vaccines
2.2. Protein-Based Vaccines
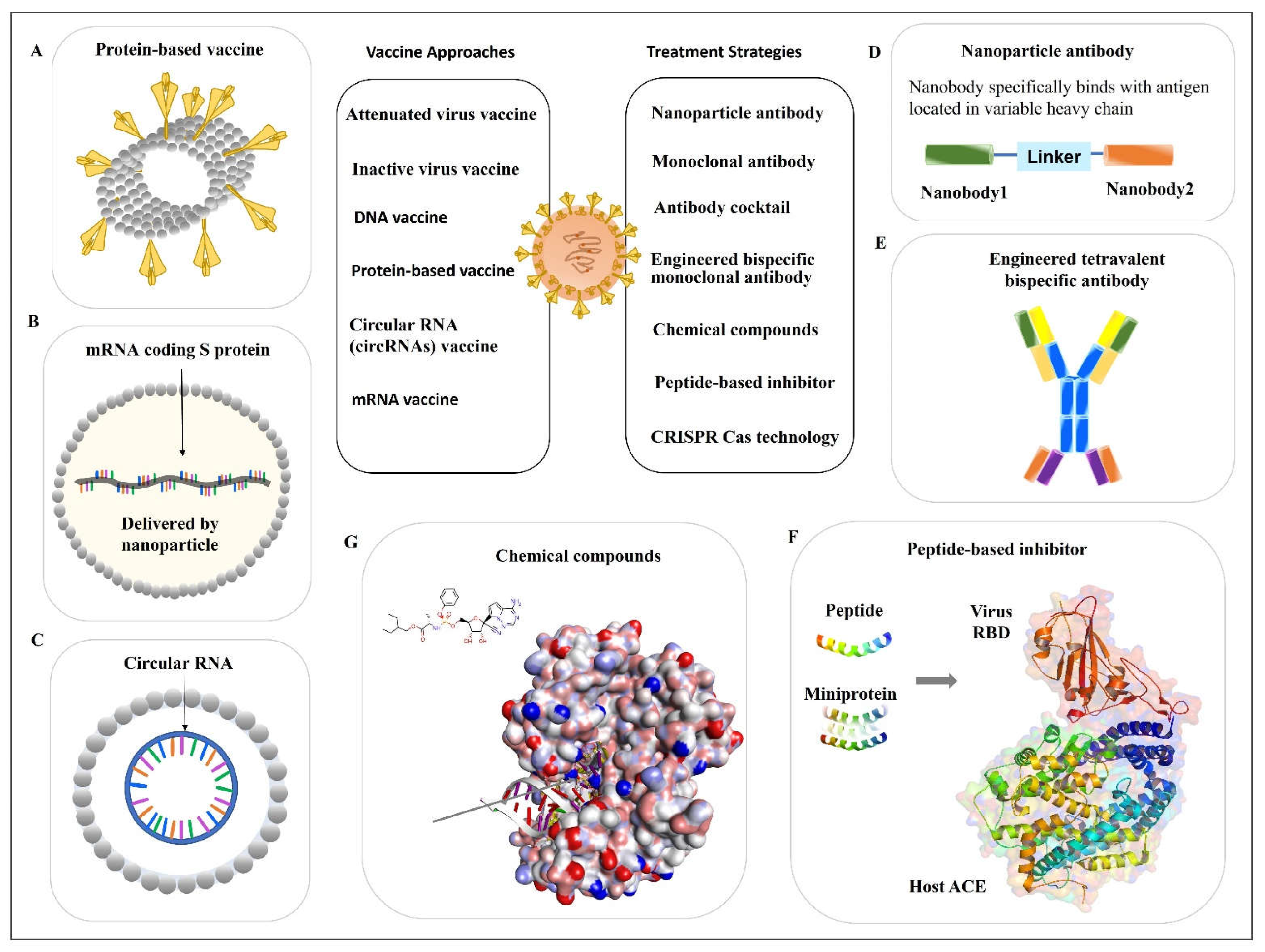
2.3. Nucleic Acid-Based Vaccines
2.3.1. DNA Vaccine
2.3.2. mRNA Vaccine
3. Antiviral Antibodies
3.1. Monoclonal Antibody
3.1.1. Monoclonal Antibody from Human Patients
3.1.2. Nanoparticle Antibody
3.2. Antibody Cocktail
3.3. Engineered Bispecific Monoclonal Antibody
4. Peptide-Based Inhibitor
This entry is adapted from the peer-reviewed paper 10.3390/ijms23116083
References
- The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544.
- Dong, E.; Du, H.; Gardner, L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020, 20, 533–534.
- Ejaz, H.; Alsrhani, A.; Zafar, A.; Javed, H.; Junaid, K.; Abdalla, A.E.; Abosalif, K.O.A.; Ahmed, Z.; Younas, S. COVID-19 and comorbidities: Deleterious impact on infected patients. J. Infect. Public Health 2020, 13, 1833–1839.
- Machhi, J.; Herskovitz, J.; Senan, A.M.; Dutta, D.; Nath, B.; Oleynikov, M.D.; Blomberg, W.R.; Meigs, D.D.; Hasan, M.; Patel, M.; et al. The Natural History, Pathobiology, and Clinical Manifestations of SARS-CoV-2 Infections. J. Neuroimmune Pharmacol. 2020, 15, 359–386.
- Yadav, R.; Chaudhary, J.K.; Jain, N.; Chaudhary, P.K.; Khanra, S.; Dhamija, P.; Sharma, A.; Kumar, A.; Handu, S. Role of Structural and Non-Structural Proteins and Therapeutic Targets of SARS-CoV-2 for COVID-19. Cells 2021, 10, 821.
- Malik, Y.A. Properties of Coronavirus and SARS-CoV-2. Malays. J. Pathol. 2020, 42, 3–11.
- Wu, C.; Liu, Y.; Yang, Y.; Zhang, P.; Zhong, W.; Wang, Y.; Wang, Q.; Xu, Y.; Li, M.; Li, X.; et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B 2020, 10, 766–788.
- Murdin, A.D.; Barreto, L.; Plotkin, S. Inactivated poliovirus vaccine: Past and present experience. Vaccine 1996, 14, 735–746.
- Vetter, V.; Denizer, G.; Friedland, L.R.; Krishnan, J.; Shapiro, M. Understanding modern-day vaccines: What you need to know. Ann. Med. 2018, 50, 110–120.
- Pollard, A.J.; Bijker, E.M. A guide to vaccinology: From basic principles to new developments. Nat. Rev. Immunol. 2021, 21, 83–100.
- Xia, S.; Zhang, Y.; Wang, Y.; Wang, H.; Yang, Y.; Gao, G.F.; Tan, W.; Wu, G.; Xu, M.; Lou, Z.; et al. Safety and immunogenicity of an inactivated SARS-CoV-2 vaccine, BBIBP-CorV: A randomised, double-blind, placebo-controlled, phase 1/2 trial. Lancet Infect. Dis. 2021, 21, 39–51.
- Hotez, P.J.; Bottazzi, M.E. Whole Inactivated Virus and Protein-Based COVID-19 Vaccines. Annu. Rev. Med. 2022, 73, 55–64.
- Li, X.N.; Huang, Y.; Wang, W.; Jing, Q.L.; Zhang, C.H.; Qin, P.Z.; Guan, W.J.; Gan, L.; Li, Y.L.; Liu, W.H.; et al. Effectiveness of inactivated SARS-CoV-2 vaccines against the Delta variant infection in Guangzhou: A test-negative case-control real-world study. Emerg. Microbes Infect. 2021, 10, 1751–1759.
- Sapkal, G.N.; Yadav, P.D.; Ella, R.; Deshpande, G.R.; Sahay, R.R.; Gupta, N.; Vadrevu, K.M.; Abraham, P.; Panda, S.; Bhargava, B. Inactivated COVID-19 vaccine BBV152/COVAXIN effectively neutralizes recently emerged B.1.1.7 variant of SARS-CoV-2. J. Travel Med. 2021, 28, taab051.
- Desai, D.; Khan, A.R.; Soneja, M.; Mittal, A.; Naik, S.; Kodan, P.; Mandal, A.; Maher, G.T.; Kumar, R.; Agarwal, A.; et al. Effectiveness of an inactivated virus-based SARS-CoV-2 vaccine, BBV152, in India: A test-negative, case-control study. Lancet Infect. Dis. 2022, 22, 349–356.
- WHO. Status of COVID-19 Vaccines within WHO EUL/PQ Evaluation Process; WHO: Geneva, Switzerland, 2021.
- Iglesias, E. Inactivated vaccines: A promising old tool against COVID-19. Res. Rev. Insights 2020, 4, 1–4.
- Iversen, P.L.; Bavari, S. Inactivated COVID-19 vaccines to make a global impact. Lancet Infect. Dis. 2021, 21, 746–748.
- Karch, C.P.; Burkhard, P. Vaccine technologies: From whole organisms to rationally designed protein assemblies. Biochem. Pharmacol. 2016, 120, 1–14.
- Tarke, A.; Coelho, C.H.; Zhang, Z.; Dan, J.M.; Yu, E.D.; Methot, N.; Bloom, N.I.; Goodwin, B.; Phillips, E.; Mallal, S.; et al. SARS-CoV-2 vaccination induces immunological T cell memory able to cross-recognize variants from Alpha to Omicron. Cell 2022, 185, 847–859.e811.
- Heath, P.T.; Galiza, E.P.; Baxter, D.N.; Boffito, M.; Browne, D.; Burns, F.; Chadwick, D.R.; Clark, R.; Cosgrove, C.; Galloway, J.; et al. Safety and Efficacy of NVX-CoV2373 COVID-19 Vaccine. N. Engl. J. Med. 2021, 385, 1172–1183.
- Chavda, V.P.; Hossain, M.K.; Beladiya, J.; Apostolopoulos, V. Nucleic Acid Vaccines for COVID-19: A Paradigm Shift in the Vaccine Development Arena. Biologics 2021, 1, 337–356.
- Voysey, M.; Clemens, S.A.C.; Madhi, S.A.; Weckx, L.Y.; Folegatti, P.M.; Aley, P.K.; Angus, B.; Baillie, V.L.; Barnabas, S.L.; Bhorat, Q.E.; et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: An interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet 2021, 397, 99–111.
- Sadoff, J.; Gray, G.; Vandebosch, A.; Cárdenas, V.; Shukarev, G.; Grinsztejn, B.; Goepfert, P.A.; Truyers, C.; Fennema, H.; Spiessens, B.; et al. Safety and Efficacy of Single-Dose Ad26.COV2.S Vaccine against COVID-19. N. Engl. J. Med. 2021, 384, 2187–2201.
- Jones, I.; Roy, P. Sputnik V COVID-19 vaccine candidate appears safe and effective. Lancet 2021, 397, 642–643.
- Karikó, K.; Buckstein, M.; Ni, H.; Weissman, D. Suppression of RNA recognition by Toll-like receptors: The impact of nucleoside modification and the evolutionary origin of RNA. Immunity 2005, 23, 165–175.
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Pérez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 mRNA COVID-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615.
- Baden, L.R.; El Sahly, H.M.; Essink, B.; Kotloff, K.; Frey, S.; Novak, R.; Diemert, D.; Spector, S.A.; Rouphael, N.; Creech, C.B.; et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021, 384, 403–416.
- Westendorf, K.; Žentelis, S.; Wang, L.; Foster, D.; Vaillancourt, P.; Wiggin, M.; Lovett, E.; van der Lee, R.; Hendle, J.; Pustilnik, A.; et al. LY-CoV1404 (bebtelovimab) potently neutralizes SARS-CoV-2 variants. bioRxiv 2022.
- Dougan, M.; Azizad, M.; Chen, P.; Feldman, B.; Frieman, M.; Igbinadolor, A.; Kumar, P.; Morris, J.; Potts, J.; Baracco, L.; et al. Bebtelovimab, alone or together with bamlanivimab and etesevimab, as a broadly neutralizing monoclonal antibody treatment for mild to moderate, ambulatory COVID-19. medRxiv 2022.
- Wu, X.; Cheng, L.; Fu, M.; Huang, B.; Zhu, L.; Xu, S.; Shi, H.; Zhang, D.; Yuan, H.; Nawaz, W.; et al. A potent bispecific nanobody protects hACE2 mice against SARS-CoV-2 infection via intranasal administration. Cell Rep. 2021, 37, 109869.
- Levin, M.J.; Ustianowski, A.; De Wit, S.; Launay, O.; Avila, M.; Templeton, A.; Yuan, Y.; Seegobin, S.; Ellery, A.; Levinson, D.J. Intramuscular AZD7442 (Tixagevimab–Cilgavimab) for Prevention of COVID-19. N. Engl. J. Med. 2022, 386.
- Levin, M.J.; Ustianowski, A.; De Wit, S.; Launay, O.; Avila, M.; Seegobin, S.; Templeton, A.; Yuan, Y.; Ambery, P.; Arends, R.H. LB5. PROVENT: Phase 3 Study of Efficacy and Safety of AZD7442 (Tixagevimab/Cilgavimab) for Pre-exposure Prophylaxis of COVID-19 in Adults. Open Forum Infect. Dis. 2021, 8, S810.
- Fenwick, C.; Turelli, P.; Ni, D.; Perez, L.; Lau, K.; Lana, E.; Pellaton, C.; Raclot, C.; Esteves-Leuenberger, L.; Campos, J. SARS-CoV-2 Omicron potently neutralized by a novel antibody with unique Spike binding properties. bioRxiv 2022.
- Su, S.-C.; Yang, T.-J.; Yu, P.-Y.; Liang, K.-H.; Chen, W.-Y.; Yang, C.-W.; Lin, H.-T.; Wang, M.-J.; Lu, R.-M.; Tso, H.-C.; et al. Structure-guided antibody cocktail for prevention and treatment of COVID-19. PLoS Pathogens 2021, 17, e1009704.
- Liang, K.-H.; Chiang, P.-Y.; Ko, S.-H.; Chou, Y.-C.; Lu, R.-M.; Lin, H.-T.; Chen, W.-Y.; Lin, Y.-L.; Tao, M.-H.; Jan, J.-T.; et al. Antibody cocktail effective against variants of SARS-CoV-2. J. Biomed. Sci. 2021, 28, 80.
- Li, C.; Zhan, W.; Yang, Z.; Tu, C.; Hu, G.; Zhang, X.; Song, W.; Du, S.; Zhu, Y.; Huang, K.; et al. Broad neutralization of SARS-CoV-2 variants by an inhalable bispecific single-domain antibody. Cell 2022, 185, 1389–1401.e1318.
- Ku, Z.; Xie, X.; Lin, J.; Gao, P.; El Sahili, A.; Su, H.; Liu, Y.; Ye, X.; Li, X.; Fan, X.; et al. Engineering SARS-CoV-2 cocktail antibodies into a bispecific format improves neutralizing potency and breadth. bioRxiv 2022.
- Zhang, C.; Yang, M. Antimicrobial Peptides: From Design to Clinical Application. Antibiotics 2022, 11, 349.
- Basit, A.; Karim, A.M.; Asif, M.; Ali, T.; Lee, J.H.; Jeon, J.H.; Rehman, S.U.; Lee, S.H. Designing Short Peptides to Block the Interaction of SARS-CoV-2 and Human ACE2 for COVID-19 Therapeutics. Front. Pharmacol. 2021, 12, 731828.
- Karoyan, P.; Vieillard, V.; Gómez-Morales, L.; Odile, E.; Guihot, A.; Luyt, C.E.; Denis, A.; Grondin, P.; Lequin, O. Human ACE2 peptide-mimics block SARS-CoV-2 pulmonary cells infection. Commun. Biol. 2021, 4, 197.
- Cao, L.; Goreshnik, I.; Coventry, B.; Case, J.B.; Miller, L.; Kozodoy, L.; Chen, R.E.; Carter, L.; Walls, A.C.; Park, Y.J.; et al. De novo design of picomolar SARS-CoV-2 miniprotein inhibitors. Science 2020, 370, 426–431.
