Epigenetic therapies describe drug molecules such as DNA methyltransferase, histone methyltransferase and histone acetylase/deacetylase inhibitors, which target epigenetic mechanisms such as DNA methylation and histone modifications. Many DNA damage response (DDR) genes are epigenetically regulated in cancer leading to transcriptional silencing and the loss of DNA repair capacity. Epigenetic marks at DDR genes, such as DNA methylation at gene promoters, have the potential to be used as stratification biomarkers, identifying which patients may benefit from particular chemotherapy treatments.
- DNA repair
- epigenetics
- cancer
- DNA methylation
1. Deoxyribonucleic Acid (DNA) Damage Response (DDR) Genes Are Epigenetically Regulated in Cancer, Affecting Chemosensitivity
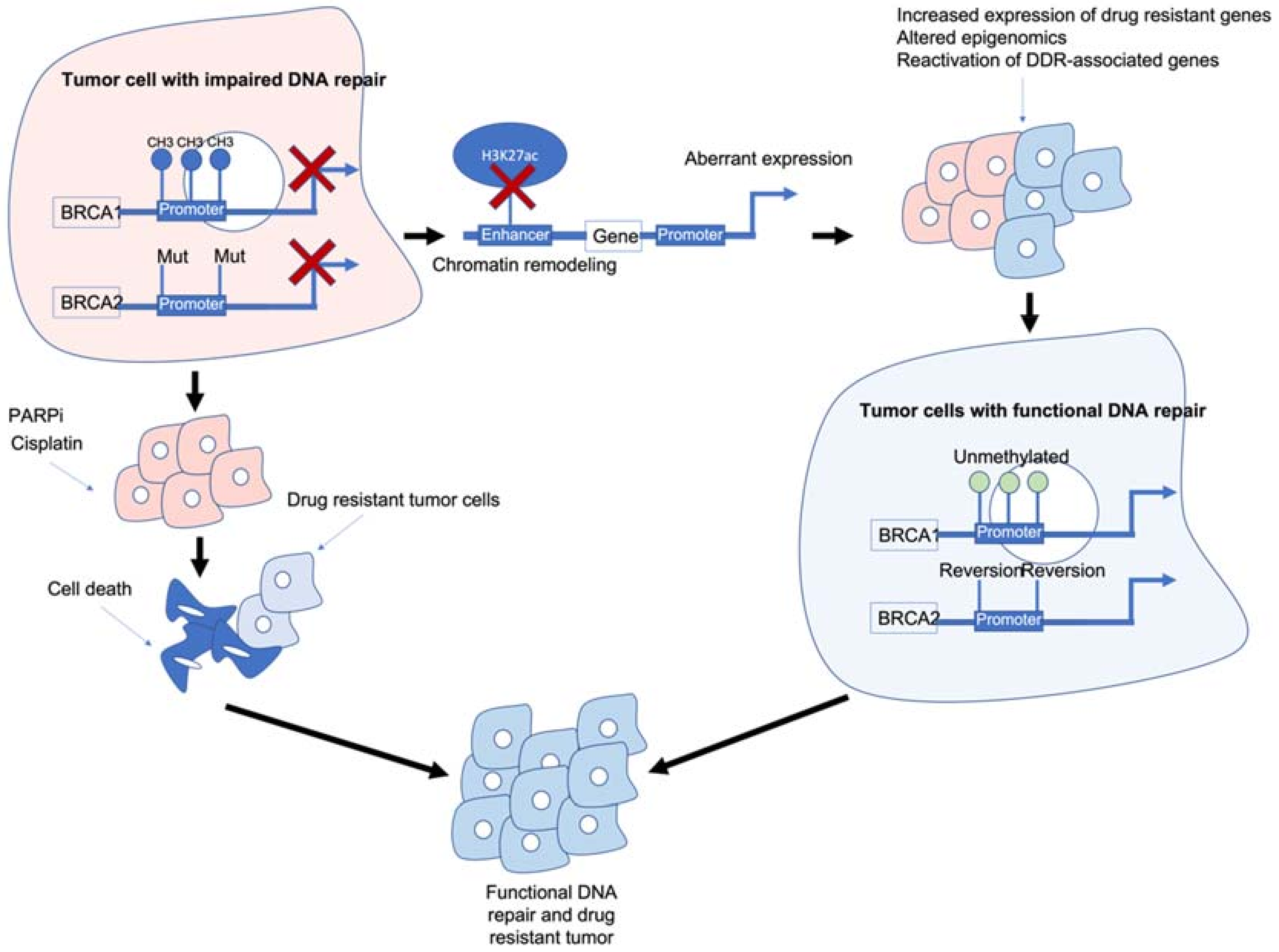 Figure 1. Epigenetic interactions with breast cancer type 1/2 susceptibility (BRCA1/2) genes and drug resistance. BRCA1/2 deficiency caused by methylation and/or mutations results in impaired DNA repair and often sensitivity to poly (ADP-ribose) polymerase inhibitors (PARPi)/platinum-based chemotherapies. BRCA1/2 deficiency can also modulate chromatin accessibility and enhancers of drug-resistant and/or DNA damage response (DDR)-associated genes. Reactivation of BRCA1/2 either by demethylation or reversion mutations can result in tumour cells with functional DDR. Ultimately, these mechanisms result in tumour cells that are drug resistant, have functional DDR and no longer respond to previous PARPi/platinum chemotherapy.
Figure 1. Epigenetic interactions with breast cancer type 1/2 susceptibility (BRCA1/2) genes and drug resistance. BRCA1/2 deficiency caused by methylation and/or mutations results in impaired DNA repair and often sensitivity to poly (ADP-ribose) polymerase inhibitors (PARPi)/platinum-based chemotherapies. BRCA1/2 deficiency can also modulate chromatin accessibility and enhancers of drug-resistant and/or DNA damage response (DDR)-associated genes. Reactivation of BRCA1/2 either by demethylation or reversion mutations can result in tumour cells with functional DDR. Ultimately, these mechanisms result in tumour cells that are drug resistant, have functional DDR and no longer respond to previous PARPi/platinum chemotherapy.2. Can Epigenetic Therapies Reverse Epigenetically Driven Drug Resistance?
| Gene | Symbol | Summary | Reference Number |
|---|---|---|---|
| O6-Methylguanine-DNA Methyltransferase | MGMT | Methylated associated with increased sensitivity to temozolomide. Enhancer region associated with increased expression and resistance to temozolomide. | [1][3][5][6][26] |
| Breast Cancer type 1 susceptibility protein | BRCA1 | Methylated associated with sensitivity to PARPi/platinum and loss of H3K27ac at enhancer regions. | [7][8][11][12][27][35] |
| Breast Cancer type 2 susceptibility protein | BRCA2 | Deficiency causes chromatin conformation changes and increased sensitivity to PARPi/platinum. | [7][8][11][12][28] |
| MutL Homolog 1 | MLH1 | Unmethylated associated with temozolomide/platinum resistance and loss of MMR. | [15][16][17][31] |
| FA Complementation Group F | FANCF | Methylation associated with sensitivity to platinum, unmethylated associated with platinum resistance. | [24][25] |
| FA Complementation Group A | FANCA | Germline mutation associated with increased sensitivity to DNA damaging agents. | [36] |
| DNA Repair Protein RAD51 homolog 1 | RAD51 | High expression associated with platinum resistance. | [37] |
| X-ray Repair Cross Complementing 2 | XRCC2 | Low expression associated with sensitivity to PARPi. | [38] |
| X-ray Repair Cross Complementing 3 | XRCC3 | Low expression associated with sensitivity to PARPi. | [38] |
| DNA Polymerase Delta 4 | POLD4 | Low expression associated with sensitivity to PARPi/platinum. | [39] |
| RAD51 Paralog C | RAD51C | Reversion mutations associated with increased resistance to PARPi. | [52] |
| RAD51 Paralog D | RAD51D | Reversion mutations associated with increased resistance to PARPi. | [52] |
| Euchromatic Histone Lysine Methyltransferase 2 | EHMT2 | Maintains repressive H3K9 methylation marks. Recruits HR-associated factors, Inhibition of EHMT2 promotes DNA damage. | [58][59][63][64][66] |
| Enhancer of Zeste Homolog 2 | EZH2 | Maintains repressive H3K27 methylation marks. Controls expression of multiple DDR-associated genes. Inhibition of EZH2 sensitises cells to PARPi. | [58][59][60][61][66] |
3. Epigenetic Changes in Normal Tissue following Chemotherapy
This entry is adapted from the peer-reviewed paper 10.3390/cancers14061533
References
- Esteller, M.; Hamilton, S.R.; Burger, P.C.; Baylin, S.B.; Herman, J.G. Inactivation of the DNA repair gene O(6)-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 1999, 59, 793–797.
- Knijnenburg, T.A.; Wang, L.; Zimmermann, M.T.; Chambwe, N.; Gao, G.F.; Cherniack, A.D.; Fan, H.; Shen, H.; Way, G.P.; Greene, C.S.; et al. Genomic and Molecular Landscape of DNA Damage Repair Deficiency across The Cancer Genome Atlas. Cell Rep. 2018, 23, 239–254.e6.
- Hegi, M.E.; Diserens, A.-C.; Gorlia, T.; Hamou, M.-F.; de Tribolet, N.; Weller, M.; Kros, J.M.; Hainfellner, J.A.; Mason, W.; Mariani, L.; et al. MGMT Gene Silencing and Benefit from Temozolomide in Glioblastoma. N. Engl. J. Med. 2005, 352, 997–1003.
- Wick, W.; Platten, M.; Meisner, C.; Felsberg, J.; Tabatabai, G.; Simon, M.; Nikkhah, G.; Papsdorf, K.; Steinbach, J.P.; Sabel, M.; et al. Temozolomide chemotherapy alone versus radiotherapy alone for malignant astrocytoma in the elderly: The NOA-08 randomised, phase 3 trial. Lancet Oncol. 2012, 13, 707–715.
- Mansouri, A.; Hachem, L.D.; Mansouri, S.; Nassiri, F.; Laperriere, N.J.; Xia, D.; Lindeman, N.I.; Wen, P.Y.; Chakravarti, A.; Mehta, M.P.; et al. MGMT promoter methylation status testing to guide therapy for glioblastoma: Refining the approach based on emerging evidence and current challenges. Neuro Oncol. 2019, 21, 167–168.
- Weller, M.; Stupp, R.; Reifenberger, G.; Brandes, A.A.; Van Den Bent, M.J.; Wick, W.; Hegi, M.E. MGMT promoter methylation in malignant gliomas: Ready for personalized medicine? Nat. Rev. Neurol. 2010, 6, 39–51.
- Bell, D.; Berchuck, A.; Birrer, M.; Chien, J.; Cramer, D.W.; Dao, F.; Dhir, R.; Disaia, P.; Gabra, H.; Glenn, P.; et al. Integrated genomic analyses of ovarian carcinoma. Nature 2011, 474, 609–615.
- Stronach, E.A.; Paul, J.; Timms, K.M.; Hughes, E.; Brown, K.; Neff, C.; Perry, M.; Gutin, A.; El-Bahrawy, M.; Steel, J.H.; et al. Biomarker assessment of HR deficiency, tumor BRCA1/2 mutations, and CCNE1 copy number in ovarian cancer: Associations with clinical outcome following platinum monotherapy. Mol. Cancer Res. 2018, 16, 1103–1111.
- Dasari, S.; Bernard Tchounwou, P. Cisplatin in cancer therapy: Molecular mechanisms of action. Eur. J. Pharmacol. 2014, 740, 364–378.
- Lord, C.J.; Ashworth, A. PARP inhibitors: Synthetic lethality in the clinic. Science 2017, 355, 1152–1158.
- Faraoni, I.; Graziani, G. Role of BRCA Mutations in Cancer Treatment with Poly(ADP-ribose) Polymerase (PARP) Inhibitors. Cancers 2018, 10, 487.
- Kondrashova, O.; Topp, M.; Nesic, K.; Lieschke, E.; Ho, G.-Y.; Harrell, M.I.; Zapparoli, G.V.; Hadley, A.; Holian, R.; Boehm, E.; et al. Methylation of all BRCA1 copies predicts response to the PARP inhibitor rucaparib in ovarian carcinoma. Nat. Commun. 2018, 9, 3970.
- Bonadio, R.; Fogace, R.; Miranda, V.; Diz, M. Homologous recombination deficiency in ovarian cancer: A review of its epidemiology and management. Clinics 2018, 73, e450s.
- Brown, R.; Curry, E.; Magnani, L.; Wilhelm-Benartzi, C.S.; Borley, J. Poised epigenetic states and acquired drug resistance in cancer. Nat. Rev. Cancer 2014, 14, 747–753.
- Wu, F.; Lu, M.; Qu, L.; Li, D.Q.; Hu, C.H. DNA methylation of hMLH1 correlates with the clinical response to cisplatin after a surgical resection in Non-small cell lung cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 5457–5463.
- Zeller, C.; Dai, W.; Steele, N.L.; Siddiq, A.; Walley, A.J.; Wilhelm-Benartzi, C.S.M.; Rizzo, S.; Van Der Zee, A.; Plumb, J.A.; Brown, R. Candidate DNA methylation drivers of acquired cisplatin resistance in ovarian cancer identified by methylome and expression profiling. Oncogene 2012, 31, 4567–4576.
- Gallitto, M.; Cheng He, R.; Inocencio, J.F.; Wang, H.; Zhang, Y.; Deikus, G.; Wasserman, I.; Strahl, M.; Smith, M.; Sebra, R.; et al. Epigenetic preconditioning with decitabine sensitizes glioblastoma to temozolomide via induction of MLH1. J. Neurooncol. 2020, 147, 557–566.
- O’Brien, V.; Brown, R. Signalling cell cycle arrest and cell death through the MMR System. Carcinogenesis 2006, 27, 682–692.
- Swisher, E.M.; Kwan, T.T.; Oza, A.M.; Tinker, A.V.; Ray-Coquard, I.; Oaknin, A.; Coleman, R.L.; Aghajanian, C.; Konecny, G.E.; O’Malley, D.M.; et al. Molecular and clinical determinants of response and resistance to rucaparib for recurrent ovarian cancer treatment in ARIEL2 (Parts 1 and 2). Nat. Commun. 2021, 12, 2487.
- Koul, S.; McKiernan, J.M.; Narayan, G.; Houldsworth, J.; Bacik, J.; Dobrzynski, D.L.; Assaad, A.M.; Mansukhani, M.; Reuter, V.E.; Bosl, G.J.; et al. Role of promoter hypermethylation in cisplatin treatment response of male germ cell tumors. Mol. Cancer 2004, 3, 16.
- Marsit, C.J.; Liu, M.; Nelson, H.H.; Posner, M.; Suzuki, M.; Kelsey, K.T. Inactivation of the Fanconi anemia/BRCA pathway in lung and oral cancers: Implications for treatment and survival. Oncogene 2004, 23, 1000–1004.
- Narayan, G.; Arias-Pulido, H.; Nandula, S.V.; Basso, K.; Sugirtharaj, D.D.; Vargas, H.; Mansukhani, M.; Villella, J.; Meyer, L.; Schneider, A.; et al. Promoter Hypermethylation of FANCF: Disruption of Fanconi Anemia-BRCA Pathway in Cervical Cancer. Cancer Res. 2004, 64, 2994–2997.
- Lim, S.L.; Smith, P.; Syed, N.; Coens, C.; Wong, H.; Van Der Burg, M.; Szlosarek, P.; Crook, T.; Green, J.A. Promoter hypermethylation of FANCF and outcome in advanced ovarian cancer. Br. J. Cancer 2008, 98, 1452–1456.
- Taniguchi, T.; Tischkowitz, M.; Ameziane, N.; Hodgson, S.V.; Mathew, C.G.; Joenje, H.; Mok, S.C.; D’Andrea, A.D. Disruption of the Fanconi anemia-BRCA pathway in cisplatin-sensitive ovarian tumors. Nat. Med. 2003, 9, 568–574.
- D’Andrea, A.D. The Fanconi Anemia/BRCA Signaling Pathway: Disruption in Cisplatin-Sensitive Ovarian Cancers. Cell Cycle 2003, 2, 289–291.
- Chen, X.; Zhang, M.; Gan, H.; Wang, H.; Lee, J.-H.; Fang, D.; Kitange, G.J.; He, L.; Hu, Z.; Parney, I.F.; et al. A novel enhancer regulates MGMT expression and promotes temozolomide resistance in glioblastoma. Nat. Commun. 2018, 9, 2949.
- Zhang, X.; Wang, Y.; Chiang, H.-C.; Hsieh, Y.-P.; Lu, C.; Park, B.H.; Jatoi, I.; Jin, V.X.; Hu, Y.; Li, R. BRCA1 mutations attenuate super-enhancer function and chromatin looping in haploinsufficient human breast epithelial cells. Breast Cancer Res. 2019, 21, 51.
- Gruber, J.J.; Chen, J.; Geller, B.; Jäger, N.; Lipchik, A.M.; Wang, G.; Kurian, A.W.; Ford, J.M.; Snyder, M.P. Chromatin Remodeling in Response to BRCA2-Crisis. Cell Rep. 2019, 28, 2182–2193.e6.
- Hung, S.; Saiakhova, A.; Faber, Z.J.; Bartels, C.F.; Neu, D.; Bayles, I.; Ojo, E.; Hong, E.S.; Pontius, W.D.; Morton, A.R.; et al. Mismatch repair-signature mutations activate gene enhancers across human colorectal cancer epigenomes. eLife 2019, 8, e40760.
- Miranda Furtado, C.L.; Dos Santos Luciano, M.C.; Da Silva Santos, R.; Furtado, G.P.; Moraes, M.O.; Pessoa, C. Epidrugs: Targeting epigenetic marks in cancer treatment. Epigenetics 2019, 14, 1164–1176.
- Plumb, J.A.; Strathdee, G.; Sludden, J.; Kaye, S.B.; Brown, R. Reversal of drug resistance in human tumor xenografts by 2′-deoxy-5-azacytidine-induced demethylation of the hMLH1 gene promoter. Cancer Res. 2000, 60, 6039–6044.
- Appleton, K.; Mackay, H.J.; Judson, I.; Plumb, J.A.; McCormick, C.; Strathdee, G.; Lee, C.; Barrett, S.; Reade, S.; Jadayel, D.; et al. Phase I and pharmacodynamic trial of the DNA methyltransferase inhibitor decitabine and carboplatin in solid tumors. J. Clin. Oncol. 2007, 25, 4603–4609.
- Glasspool, R.M.; Brown, R.; Gore, M.E.; Rustin, G.J.S.; McNeish, I.A.; Wilson, R.H.; Pledge, S.; Paul, J.; MacKean, M.; Hall, G.D.; et al. A randomised, phase II trial of the DNA-hypomethylating agent 5-aza-2′-deoxycytidine (decitabine) in combination with carboplatin vs carboplatin alone in patients with recurrent, partially platinum-sensitive ovarian cancer. Br. J. Cancer 2014, 110, 1923–1929.
- Oza, A.M.; Matulonis, U.A.; Alvarez Secord, A.; Nemunaitis, J.; Roman, L.D.; Blagden, S.P.; Banerjee, S.; McGuire, W.P.; Ghamande, S.; Birrer, M.J.; et al. A Randomized Phase II Trial of Epigenetic Priming with Guadecitabine and Carboplatin in Platinum-resistant, Recurrent Ovarian Cancer. Clin. Cancer Res. 2020, 26, 1009–1016.
- Bouwman, P.; Jonkers, J. Molecular pathways: How can BRCA-mutated tumors become resistant to PARP inhibitors? Clin. Cancer Res. 2014, 20, 540–547.
- Wilkes, D.C.; Sailer, V.; Xue, H.; Cheng, H.; Collins, C.C.; Gleave, M.; Wang, Y.; Demichelis, F.; Beltran, H.; Rubin, M.A.; et al. A germline FANCA alteration that is associated with increased sensitivity to DNA damaging agents. Mol. Case Stud. 2017, 3, a001487.
- Hoppe, M.M.; Jaynes, P.; Wardyn, J.D.; Upadhyayula, S.S.; Tan, T.Z.; Lie, S.; Lim, D.G.Z.; Pang, B.N.K.; Lim, S.P.S.; Yeong, J.; et al. Quantitative imaging of RAD51 expression as a marker of platinum resistance in ovarian cancer. EMBO Mol. Med. 2021, 13, e13366.
- Murai, J.; Huang, S.Y.N.; Das, B.B.; Renaud, A.; Zhang, Y.; Doroshow, J.H.; Ji, J.; Takeda, S.; Pommier, Y. Trapping of PARP1 and PARP2 by clinical PARP inhibitors. Cancer Res. 2012, 72, 5588–5599.
- Zhang, S.; Chao, H.H.; Wang, X.; Zhang, Z.; Lee, E.Y.C.; Lee, M.Y.W.T. Loss of the p12 subunit of DNA polymerase delta leads to a defect in HR and sensitization to PARP inhibitors. DNA Repair 2019, 73, 64–70.
- Ledermann, J.A.; El-Khouly, F. PARP inhibitors in ovarian cancer: Clinical evidence for informed treatment decisions. Br. J. Cancer 2015, 113, S10–S16.
- Domchek, S.M.; Aghajanian, C.; Shapira-Frommer, R.; Schmutzler, R.K.; Audeh, M.W.; Friedlander, M.; Balmaña, J.; Mitchell, G.; Fried, G.; Stemmer, S.M.; et al. Efficacy and safety of olaparib monotherapy in germline BRCA1/2 mutation carriers with advanced ovarian cancer and three or more lines of prior therapy. Gynecol. Oncol. 2016, 140, 199–203.
- Matulonis, U.A.; Penson, R.T.; Domchek, S.M.; Kaufman, B.; Shapira-Frommer, R.; Audeh, M.W.; Kaye, S.; Molife, L.R.; Gelmon, K.A.; Robertson, J.D.; et al. Olaparib monotherapy in patients with advanced relapsed ovarian cancer and a germline BRCA1/2 mutation: A multistudy analysis of response rates and safety. Ann. Oncol. 2016, 27, 1013–1019.
- Kim, D.S.; Camacho, C.V.; Nagari, A.; Malladi, V.S.; Challa, S.; Kraus, W.L. Activation of PARP-1 by snoRNAs Controls Ribosome Biogenesis and Cell Growth via the RNA Helicase DDX21. Mol. Cell 2019, 75, 1270–1285.e14.
- Keung, M.Y.; Wu, Y.; Badar, F.; Vadgama, J.V. Response of Breast Cancer Cells to PARP Inhibitors Is Independent of BRCA Status. J. Clin. Med. 2020, 9, 940.
- Luijsterburg, M.S.; de Krijger, I.; Wiegant, W.W.; Shah, R.G.; Smeenk, G.; de Groot, A.J.L.; Pines, A.; Vertegaal, A.C.O.; Jacobs, J.J.L.; Shah, G.M.; et al. PARP1 Links CHD2-Mediated Chromatin Expansion and H3.3 Deposition to DNA Repair by Non-homologous End-Joining. Mol. Cell 2016, 61, 547–562.
- Audeh, M.W.; Carmichael, J.; Penson, R.T.; Friedlander, M.; Powell, B.; Bell-McGuinn, K.M.; Scott, C.; Weitzel, J.N.; Oaknin, A.; Loman, N.; et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: A proof-of-concept trial. Lancet 2010, 376, 245–251.
- Sonnenblick, A.; de Azambuja, E.; Azim, H.A.; Piccart, M. An update on PARP inhibitors—Moving to the adjuvant setting. Nat. Rev. Clin. Oncol. 2015, 12, 27–41.
- Sakai, W.; Swisher, E.M.; Jacquemont, C.; Chandramohan, K.V.; Couch, F.J.; Langdon, S.P.; Wurz, K.; Higgins, J.; Villegas, E.; Taniguchi, T. Functional restoration of BRCA2 protein by secondary BRCA2 mutations in BRCA2-mutated ovarian carcinoma. Cancer Res. 2009, 69, 6381–6386.
- Dhillon, K.K.; Swisher, E.M.; Taniguchi, T. Secondary mutations of BRCA1/2 and drug resistance. Cancer Sci. 2011, 102, 663–669.
- Norquist, B.; Wurz, K.A.; Pennil, C.C.; Garcia, R.; Gross, J.; Sakai, W.; Karlan, B.Y.; Taniguchi, T.; Swisher, E.M. Secondary Somatic Mutations Restoring BRCA1/2 Predict Chemotherapy Resistance in Hereditary Ovarian Carcinomas. J. Clin. Oncol. 2011, 29, 3008–3015.
- Lin, K.K.; Harrell, M.I.; Oza, A.M.; Oaknin, A.; Ray-Coquard, I.; Tinker, A.V.; Helman, E.; Radke, M.R.; Say, C.; Vo, L.T.; et al. BRCA Reversion Mutations in Circulating Tumor DNA Predict Primary and Acquired Resistance to the PARP Inhibitor Rucaparib in High-Grade Ovarian Carcinoma. Cancer Discov. 2019, 9, 210–219.
- Kondrashova, O.; Nguyen, M.; Shield-Artin, K.; Tinker, A.V.; Teng, N.N.H.; Harrell, M.I.; Kuiper, M.J.; Ho, G.Y.; Barker, H.; Jasin, M.; et al. Secondary somatic mutations restoring RAD51C and RAD51D associated with acquired resistance to the PARP inhibitor rucaparib in high-grade ovarian carcinoma. Cancer Discov. 2017, 7, 984–998.
- Curry, E.; Zeller, C.; Masrour, N.; Patten, D.K.; Gallon, J.; Wilhelm-Benartzi, C.S.; Ghaem-Maghami, S.; Bowtell, D.D.; Brown, R. Genes predisposed to DNA hypermethylation during acquired resistance to chemotherapy are identified in ovarian tumors by bivalent chromatin domains at initial diagnosis. Cancer Res. 2018, 78, 1383–1391.
- Wei, S.; Li, C.; Yin, Z.; Wen, J.; Meng, H.; Xue, L.; Wang, J. Histone methylation in DNA repair and clinical practice: New findings during the past 5-years. J. Cancer 2018, 9, 2072–2081.
- Sun, J.; Cai, X.; Yung, M.M.; Zhou, W.; Li, J.; Zhang, Y.; Li, Z.; Liu, S.S.; Cheung, A.N.Y.; Ngan, H.Y.S.; et al. miR-137 mediates the functional link between c-Myc and EZH2 that regulates cisplatin resistance in ovarian cancer. Oncogene 2019, 38, 564–580.
- Duan, R.; Du, W.; Guo, W. EZH2: A novel target for cancer treatment. J. Hematol. Oncol. 2020, 13, 104.
- Andronikou, C.; Rottenberg, S. Studying PAR-Dependent Chromatin Remodeling to Tackle PARPi Resistance. Trends Mol. Med. 2021, 27, 630–642.
- Yamaguchi, H.; Du, Y.; Nakai, K.; Ding, M.; Chang, S.S.; Hsu, J.L.; Yao, J.; Wei, Y.; Nie, L.; Jiao, S.; et al. EZH2 contributes to the response to PARP inhibitors through its PARP-mediated poly-ADP ribosylation in breast cancer. Oncogene 2018, 37, 208–217.
- Rao, Z.Y.; Cai, M.Y.; Yang, G.F.; He, L.R.; Mai, S.J.; Hua, W.F.; Liao, Y.J.; Deng, H.X.; Chen, Y.C.; Guan, X.Y.; et al. EZH2 supports ovarian carcinoma cell invasion and/or metastasis via regulation of TGF-β 1 and is a predictor of outcome in ovarian carcinoma patients. Carcinogenesis 2010, 31, 1576–1583.
- Yang, Y.A.; Yu, J. EZH2, an epigenetic driver of prostate cancer. Protein Cell 2013, 4, 331–341.
- Karakashev, S.; Fukumoto, T.; Zhao, B.; Lin, J.; Wu, S.; Fatkhutdinov, N.; Park, P.H.; Semenova, G.; Jean, S.; Cadungog, M.G.; et al. EZH2 Inhibition Sensitizes CARM1-High, Homologous Recombination Proficient Ovarian Cancers to PARP Inhibition. Cancer Cell 2020, 37, 157–167.e6.
- Caruso, L.B.; Martin, K.A.; Lauretti, E.; Hulse, M.; Siciliano, M.; Lupey-Green, L.N.; Abraham, A.; Skorski, T.; Tempera, I. Poly(ADP-ribose) Polymerase 1, PARP1, modifies EZH2 and inhibits EZH2 histone methyltransferase activity after DNA damage. Oncotarget 2018, 9, 10585–10605.
- Yang, Q.; Zhu, Q.; Lu, X.; Du, Y.; Cao, L.; Shen, C.; Hou, T.; Li, M.; Li, Z.; Liu, C.; et al. G9a coordinates with the RPA complex to promote DNA damage repair and cell survival. Proc. Natl. Acad. Sci. USA 2017, 114, E6054–E6063.
- Watson, Z.L.; Yamamoto, T.M.; McMellen, A.; Kim, H.; Hughes, C.J.; Wheeler, L.J.; Post, M.D.; Behbakht, K.; Bitler, B.G. Histone methyltransferases EHMT1 and EHMT2 (GLP/G9A) maintain PARP inhibitor resistance in high-grade serous ovarian carcinoma. Clin. Epigenet. 2019, 11, 165.
- Chen, M. Efficacy of PARP inhibition combined with EZH2 inhibition depends on BRCA mutation status and microenvironment in breast cancer. FEBS J. 2021, 288, 2884–2887.
- Curry, E.; Green, I.; Chapman-Rothe, N.; Shamsaei, E.; Kandil, S.; Cherblanc, F.L.; Payne, L.; Bell, E.; Ganesh, T.; Srimongkolpithak, N.; et al. Dual EZH2 and EHMT2 histone methyltransferase inhibition increases biological efficacy in breast cancer cells. Clin. Epigenet. 2015, 7, 84.
- Li, G.-M. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008, 18, 85–98.
- O’Hagan, H.M.; Wang, W.; Sen, S.; DeStefano Shields, C.; Lee, S.S.; Zhang, Y.W.; Clements, E.G.; Cai, Y.; Van Neste, L.; Easwaran, H.; et al. Oxidative Damage Targets Complexes Containing DNA Methyltransferases, SIRT1, and Polycomb Members to Promoter CpG Islands. Cancer Cell 2011, 20, 606–619.
- O’Hagan, H.M.; Mohammad, H.P.; Baylin, S.B. Double Strand Breaks Can Initiate Gene Silencing and SIRT1-Dependent Onset of DNA Methylation in an Exogenous Promoter CpG Island. PLoS Genet. 2008, 4, e1000155.
- Ding, N.; Bonham, E.M.; Hannon, B.E.; Amick, T.R.; Baylin, S.B.; O’Hagan, H.M. Mismatch repair proteins recruit DNA methyltransferase 1 to sites of oxidative DNA damage. J. Mol. Cell Biol. 2016, 8, 244–254.
- Cuozzo, C.; Porcellini, A.; Angrisano, T.; Morano, A.; Lee, B.; Di Pardo, A.; Messina, S.; Iuliano, R.; Fusco, A.; Santillo, M.R.; et al. DNA Damage, Homology-Directed Repair, and DNA Methylation. PLoS Genet. 2007, 3, e110.
- Flanagan, J.M.; Wilson, A.; Koo, C.; Masrour, N.; Gallon, J.; Loomis, E.; Flower, K.; Wilhelm-Benartzi, C.; Hergovich, A.; Cunnea, P.; et al. Platinum-based chemotherapy induces methylation changes in blood DNA associated with overall survival in patients with ovarian cancer. Clin. Cancer Res. 2017, 23, 2213–2222.
- Shenker, N.S.; Polidoro, S.; van Veldhoven, K.; Sacerdote, C.; Ricceri, F.; Birrell, M.A.; Belvisi, M.G.; Brown, R.; Vineis, P.; Flanagan, J.M. Epigenome-wide association study in the European Prospective Investigation Into Cancer And Nutrition (EPIC-Turin) identifies novel genetic loci associated with smoking. Hum. Mol. Genet. 2013, 22, 843–851.
- Flanagan, J.M.; Brook, M.N.; Orr, N.; Tomczyk, K.; Coulson, P.; Fletcher, O.; Jones, M.E.; Schoemaker, M.J.; Ashworth, A.; Swerdlow, A.; et al. Temporal stability and determinants of white blood cell DNA methylation in the breakthrough generations study. Cancer Epidemiol. Biomark. Prev. 2015, 24, 221–229.
- Fasanelli, F.; Baglietto, L.; Ponzi, E.; Guida, F.; Campanella, G.; Johansson, M.; Grankvist, K.; Johansson, M.; Assumma, M.B.; Naccarati, A.; et al. Hypomethylation of smoking-related genes is associated with future lung cancer in four prospective cohorts. Nat. Commun. 2015, 6, 10192.
- Rumgay, H.; Shield, K.; Charvat, H.; Ferrari, P.; Sornpaisarn, B.; Obot, I.; Islami, F.; Lemmens, V.E.P.P.; Rehm, J.; Soerjomataram, I. Global burden of cancer in 2020 attributable to alcohol consumption: A population-based study. Lancet Oncol. 2021, 22, 1071–1080.
- Liu, C.; Marioni, R.E.; Hedman, A.K.; Pfeiffer, L.; Tsai, P.C.; Reynolds, L.M.; Just, A.C.; Duan, Q.; Boer, C.G.; Tanaka, T.; et al. A DNA methylation biomarker of alcohol consumption. Mol. Psychiatry 2018, 23, 422–433.
- Jones, P.A.; Ohtani, H.; Chakravarthy, A.; De Carvalho, D.D. Epigenetic therapy in immune-oncology. Nat. Rev. Cancer 2019, 19, 151–161.
