α-Lactalbumin (α-LA) is a small (Mr 14,200), acidic (pI 4-5), Ca2+-binding protein. α-LA is a regulatory component of lactose synthase enzyme system. α-LA is very important in infant nutrition since it constitutes a large part of the whey and total protein in human milk. The protein possesses a single strong Ca2+-binding site, which can also bind Mg2+, Mn2+, Na+, K+, and some other metal cations. It contains several distinct Zn2+-binding sites. Physical properties of α-LA strongly depend on the occupation of its metal binding sites by metal ions. In the absence of bound metal ions α-LA is in the molten globule-like state. The binding of metal ions, and especially of Ca2+, increases stability of α-LA against action of heat, various denaturing agents, and proteases, while the binding of Zn2+ to the Ca2+-loaded protein decreases its stability and causes its aggregation. The thermal unfolding of apo-α-LA takes place in the temperature region from 10 to 30 °C. The binding of Ca2+ under the conditions of low ionic strength shifts the thermal transition to higher temperatures by more than 40 °C. The binding of Mg2+, Na+, and K+ increases protein stability as well. The stronger an ion binds to the protein, the more pronounced the thermal transition shift. All four classes of surfactants (anionic, cationic, non-ionic, and zwitterionic) denature α-LA and the denaturation involves at least one intermediate. The position of any denaturation transition in α-LA (half-transition temperature, half-transition pressure, half-transition denaturant concentration) depends upon metal ion concentration in solution (especially if this metal ion is Ca2+). Therefore, values of denaturation temperature or urea or guanidine hydrochloride denaturing concentration are relatively meaningless for α-LA without specifying the metal ion content(s) and their solution concentration(s). At a neutral or slightly acidic pH at a physiological temperature, α-LA can associate with membranes. The conformations of the membrane-bound protein range from native-like to molten globule-like states. At a low pH, α-LA penetrates the interior of the negatively charged membranes and exhibits a molten globule conformation. Depending on external conditions, α-LA can form amyloid fibrils, amorphous aggregates, nanoparticles, and nanotubes. At pH 2, α-LA in the classical molten globule conformation can form amyloid fibrils. Some of these aggregated states of α-LA (nanoparticles, nanotubes) can be used in practical applications such as drug delivery to tissues and organs. The structure and self-assembly behavior of α-LA are governed by a subtle balance between hydrophobic and polar interactions and this balance can be finely tuned through the addition of selected substances. Small size nanoparticles of α-LA (100 to 200 nm) can be obtained with the use of various desolvating agents. Partially hydrolyzed α-LA can form nanotubes. α-LA and some of its fragments possess bactericidal and antiviral activities. Complexes of partially unfolded α-LA with oleic acid showed significant cytotoxicity to various tumor and bacterial cells. α-LA in such complexes plays a role of a delivery carrier of cytotoxic fatty acid molecules into tumor cells across the cell membrane. Cytotoxic protein–oleic acid complexes possess a common core-shell structure, where an oily core is made of a micellar oleic acid, whereas a proteinaceous shell, which stabilizes the oleic acid micelle, is formed from the flexible, partially unfolded proteins. These complexes called liprotides (lipids and partially denatured proteins), which are potential novel anti-tumorous drugs, can be considered as molten globular containers filled with the toxic oil.
- alpha-lactalbumin
- structure
- calcium binding
- folding
- molten globule
- cytotoxicity
1. Introduction
α-Lactalbumin (α-LA) is a small (Mr 14,200), acidic (pI 4–5) globular protein found in the whey fraction of milk in all mammals. Mean concentration of α-LA in mature human milk is 2.44 ± 0.64 g/L during lactation [1]. α-LA accounts for about 25–35% of the total milk protein content and 41% of the whey protein content [2][3]. α-LA is one of the two components of lactose synthase, which catalyzes the final step in lactose biosynthesis in the lactating mammary gland [4]. The second component of this system is galactosyl transferase (GT), which is involved in the processing of proteins in various secretory cells by transferring galactosyl groups from UDP-galactose to glycoproteins containing N-acetylglucosamine. In the lactating mammary gland the specificity of GT is modulated by its interaction with α-LA, which increases its affinity and specificity for glucose.
α-LA has a single strong Ca2+-binding site [5][6] and for this reason it frequently serves as a simple model Ca2+-binding protein. α-LA has several partially folded intermediate states, which have been of intense interest in the general protein folding field. Both its acidic pH state and metal free state (at elevated temperatures) are classical “molten globule” [7][8]. Some forms of α-LA can induce apoptosis in tumor cells [9] and possess bactericidal activity [10]. Depending on conditions, α-LA can form fibrils, nanoparticles, and nanotubes. It is an amazing protein and studies of this protein have been going on for several decades and it does not look like they will end soon. Several reviews on α-LA were published in 2000 [11], 2005 [12], 2009 [13], 2011 [14], 2016 [15][16][17] and 2020 [18].
2. α-Lactalbumin Structure
α-Lactalbumins (α-LA), lysozymes c, and calcium-binding lysozymes make up a protein super-family [19][20][21][22]. Their genes consist of four exons separated by three introns [23][24][25]. α-LA gene is expressed in epithelial cells of the lactating mammary gland. Specific amino acid substitutions in α-LA resulted in the loss of the lysozyme catalytic activity and created the features necessary for the role of α-LA in lactose synthesis. Most α-LAs consist of 123 amino acid residues, only rat α-LA contains 17 additional C-terminal residues. All α-LAs contain 8 cysteines, which form four disulfide bonds crucial for the formation of the native fold of these proteins [26]. The UniProt database (http://www.uniprot.org/uniprot/) contains the amino acid sequences of α-LAs from 22 species. Positions of all their eight cysteines and a calcium-binding site (residues DKFLDDDITDDI in human protein) are strongly conserved.
Usually, a part of α-LA is glycosylated in fresh milk [27]. Rat α-LA is mostly glycosylated (mannose, galactose, glucose, glucosamine, galactosamine, fucose, and sialic acid) and exists in multiple forms [28].
The amino acid compositions of α-LAs are generally closer to the compositions of ordered proteins than to the compositions of intrinsically disordered proteins [15]. The intrinsic disorder propensities of α-LAs from various species evaluated by several disorder predictors showed that α-LAs are expected to be compact ordered proteins containing functionally important short regions of intrinsic disorder [15]. In various α-LAs, a part of the region corresponding to the Ca2+-binding motif is predicted to be either disordered (disorder scores above 0.5) or at least flexible having disorder scores exceeding noticeably 0.1 [15].
X-ray crystallographic data are available for α-LAs isolated from human (PDB ID: 1A4V), baboon (PDB ID: 1ALC), cow (PDB ID: 1F6R), goat (PDB ID: 1HFY), and guinea pig (PDB ID: 1HFX) milk. In fact, the overall structures of guinea-pig [29], goat [29][30], bovine [29], buffalo [31], and human [32] α-LAs are similar to the structure of that earlier reported for baboon α-LA [33]. X-ray crystallography showed that the 3D structure of α-LA is very similar to that of lysozyme [32][33]. The α-LA molecule is roughly ellipsoidal in shape (23 × 26 × 40 Å) and consists of two domains: A large α-helical domain and a small mostly β-structural domain connected by a calcium-binding loop (Figure 1). The α-helical domain is composed of three classical α-helices (residues 5–11 (A), 23–34 (B), and 86–99 (C)) and two short 310-helices (residues 17–21 and 115–119). The small β-sheet domain is composed of a series of loops, a small three-stranded antiparallel β-pleated sheet (residues 40–43, 47–50, and 55–56) and a short 310-helix (76–82). The two domains are separated by a deep cleft between them. At the same time, they are held together by the cysteine bridges between residues 73 and 91 and between residues 61 and 77. The major secondary structural elements are conserved in all the α-LA structures.
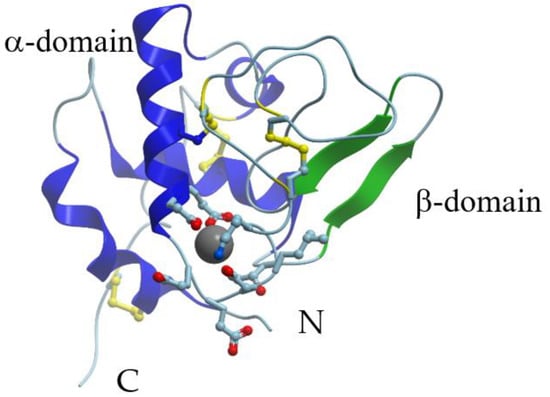
Figure 1. X-ray structure of Ca2+-bound human α-lactalbumin (PDB ID 1a4v). α-Helices are shown in blue; β-structure is shown in green; S-S bridges are shown in yellow. Gray sphere represents bound calcium ion, and amino acid residues taking part in its coordination are represented by stick model.
At low pH values (pH 2) α-LA is in the classical molten globule state (A-state) [7][34]. This state has a relatively high content of native-like secondary structure but its tertiary structure is partially disordered and many amino acid side chains are not entirely fixed and characterized by an elevated flexibility. The radius of gyration of native Ca2+-loaded α-LA is 15.7 Å, but the acid molten globule has a radius of 17.2 Å [35]. The similar partially folded, molten globule state of α-LA is formed in the calcium-depleted form at neutral pH upon moderate heating by dissolving the protein in aqueous trifluoroethanol, by adding fatty acid [36], or in the course of urea- and guanidine chloride-induced protein unfolding [37][38]. Most of the aromatic amino acid residues in α-LA molten globule are more accessible to solvent than in the native state [39].
The formation of disulfide bridges in α-LA is extremely important for its correct folding [26]. At the same time, an α-LA mutant, in which all eight cysteines were substituted by alanine, was nearly as compact as wild-type α-LA at an acidic pH [40]. It shows that overall compaction of the α-LA fold is determined by the polypeptide sequence itself and does not represent a result of the disulfide bond cross-linking.
α-LA has a single strong calcium-binding site, which is formed by oxygen ligands from carboxylic groups of three Asp residues (82, 87, and 88) and from two carbonyl groups of the peptide backbone (Lys79 and Asp84) in a loop between two helices [32][33]. In addition, one or two water molecules take part in direct coordination of Ca2+. Overall, the oxygen ligands form a distorted pentagonal bipyramidal structure. The binding of Ca2+ to α-LA at room and elevated temperatures causes pronounced changes mostly in the tertiary structure of this protein, whereas its secondary structure is almost unaffected by Ca2+ binding or release.
The thermal unfolding of apo-α-LA takes place in the temperature region from 10 to 30 °C. The binding of calcium and other metal ions shifts the thermal transition toward higher temperatures. The magnitude of the shift is highly dependent on the type of bound metal cation. The equilibrium scheme of the binding of one metal ion to the protein molecule should take into consideration equilibria between native and thermally changed states of the protein [37][41]. It was found that Ca2+ and Mg2+ cations compete with each other for the same site in α-LA [42]. The kinetics of Mg2+ association with α-LA is about four orders of magnitude slower compared to the kinetics of Ca2+ binding, whereas the dissociation of both cations is equally slow [43]. The tight association of water molecules to free Mg2+ ions, which decreases the entropy of the water–metal system, is probably responsible for higher entropy changes upon Mg2+ binding to protein.
Several distinct Zn2+-binding sites were found in α-LA [44][45]. One zinc ion is sandwiched between Glu49 and Glu116 of the symmetry-related subunit in the dimeric crystal unit cell of human α-LA [46]. However, the strongest zinc-binding site appears to be located near the N-terminus of the protein [47]. A proposed site consists of oxygens from Glu1, Glu7, Glu11, and Asp37.
3. Folding-Unfolding of α-Lactalbumin
The folding process of α-LA was studied by many experimental methods. Ity was found that the folding nucleus of goat α-LA is located near the Ca2+-binding site and at the interface between the C-helix and the β domain [48]. The radius of gyration and the overall shape of the kinetic folding intermediate of α-LA are the same as those of the molten globule state observed at equilibrium. The folding intermediate is more hydrated than the native state and the hydrated water molecules go away during the transition from the molten globule to the native state [49]. Upon the binding of Ca2+ to the molten globule state of α-LA, the calcium-binding site acts as a nucleus for the stabilization of the tertiary structure in the rest of the protein [50]. A neutron-scattering study of the nanosecond and picosecond dynamics of the native and denatured α-LA revealed that under extremely denaturing conditions and even in the absence of disulfide bonds α-LA possesses some α-helical structure and tertiary-like side-chain interactions fluctuating on sub-nanosecond time-scales [51].
All four classes of surfactants (anionic, cationic, non-ionic, and zwitterionic) denature α-LA and the denaturation involves at least one intermediate [52]. Non-ionic and zwitterionic surfactant monomers are able to prepare α-LA for denaturation due to weak monomer–protein interactions that facilitate micellar interactions, while ionic surfactants denature α-LA efficiently as monomers/hemi-micellar aggregates below the critical micelle concentration (cmc) but exclusively as micelles above the cmc. The binding of metal cations seriously increases the stability of α-LA not only against heat, but also against the action of various denaturing agents such as urea or GdnHCl [37]. Important features of the urea or GdnHCl denaturation curves are distinct intermediate molten globule-like states arising at intermediate denaturant concentrations. It was found that heat denatured proteins contain secondary structure, and GdnHCl (or urea) induces a cooperative transition between heat-denatured and GdnHCl-denatured states [53].
4. Fibrillation of α-Lactalbumin; Nanoparticles and Nanotubes
Protein misfolding and subsequent formation of protein aggregates of various morphologies is the cause of many diseases. It was found that at pH 2 bovine α-LA in the classical molten globule conformation can form amyloid fibrils [54]. S-(carboxymethyl)-α-LA, a disordered form of the protein with three out of four disulfide bridges reduced, was even more susceptible to fibrillation. The fibrillation was accompanied by a dramatic increase in the β-structure content monitored by FTIR spectroscopy and by a characteristic increase in the thioflavin T fluorescence intensity. α-LA derivatives with a single peptide bond fission (1–40/41–123, named Th1–α-LA) or a deletion of a chain segment of twelve amino acid residues located at the level of the β-domain of the native protein (1–40/53–123, named desβ–α-LA) aggregate at pH 2.0 much faster than the intact protein and form long and well-ordered fibrils [55]. In contrast to intact α-LA, the α-LA derivatives form regular fibrils also at a neutral pH. The structure and self-assembly behavior of α-LA are governed by a subtle balance between hydrophobic and polar interactions and this balance can be finely tuned through the addition of selected hydrophobic monohydric alcohols [56].
Small size nanoparticles of α-LA (100 to 200 nm) can be obtained with the use of acetone as the desolvating agent and without any pretreatment [57]. These nanoparticles, with an isoelectric point of 3.61, are very stable at pH values >4.8, although their antioxidant activity is weak. Zhang et al. prepared silver/protein nanocomposites using α-LA [58]. The nanocomposites were formed by Ag–S or Ag–N coordination bonds and electrostatic interactions.
α-LA is characterized by the ability to form fibrillar nanostructures called “nanotubes”. For example, partially hydrolyzed α-LA forms strong gels consisting of non-branching, hollow microtubules with a uniform diameter distribution of about 20 nm and lengths exceeding 2 μm [59]. Formation of nanotubes from partially hydrolyzed α-LA was investigated at various pH values, two concentrations of α-LA, and two calcium levels [60]. Nanotubes were formed under almost all combinations of the investigated factors. Only one sample (10 g L−1, calcium ratio 2.4, pH 7.5) formed mainly fibrils instead of nanotubes. The majority of nanotubes were found to have an outer diameter around 19 and an inner diameter of 6.6 nm.
5. Interactions of α-Lactalbumin with Organic Substances, Peptides, and Proteins
α-LA can interact with various organic substances and the interaction depends on calcium and other physiologically significant metal ions. α-LA binds UDP-galactose, the substrate of lactose synthase reaction, as well as UDP and UTP [61]. The binding parameters depend upon the metal bound state of the protein, but the binding constant for UDP-galactose is low (103 − 104 M−1).
The binding of bis-ANS (hydrophobic fluorescent probe 1,1’-bis(4-anilino-5-naphthalene sulfonate) is widely used for studies of α-LA [62]. Apo-α-LA binds two bis-ANS molecules per molecule, while Ca2+-α-LA binds five bis-ANS molecules. ANS binds to the acidic state of α-LA at two independent binding sites, which remain nearly the same in the temperature range of 10–35 °C [63].
α-LA binds melittin, a short peptide from bee venom, which is frequently used as a model target protein for calmodulin and other calcium-binding proteins [64]. Calmodulin and some other calcium-binding proteins bind melittin mostly in the presence of Ca2+. In contrast, α-LA is able to bind melittin only in the absence of calcium ions. Apo-protein interacts with melittin with the binding constant 5 × 107 M−1. The binding alters the melittin conformation from a random coil in solution to a helical structure in the binary complex with apo-α-LA.
α-LA possesses several classes of fatty acid-binding sites [65][66]. Using fluorescence spectroscopy it was shown that bovine apo-α-LA has one binding site for stearic acid with a dissociation constant of 2.3 µM at pH 8.5, whereas several binding sites (n = 3 − 5) with a dissociation constant of 35 µM were found for 5-doxyl stearic acid [66].
Interestingly, α-LA interacts with lysozyme. These two globular proteins have highly homologous tertiary structures but opposite electric charges. As assessed by isothermal titration calorimetry, lysozyme does not bind to the Ca2+-loaded α-LA, but interacts with calcium-depleted α-LA [67][68]. This interaction leads to the formation of various supramolecular structures depending on the temperature.
6. Interactions of α-Lactalbumin with Membranes
α-LA interacts with membranes, despite the fact that it is a soluble protein. This interaction is governed by the charge of the lipid headgroup, membrane curvature, composition, ionic strength, and pH. The self-incorporation of apo-α-LA into single unilamellar vesicles (SUV) of dimyristoylphosphatidylcholine (DMPC) and dipalmitoylphosphatidylcholine (DPPC) was demonstrated by size exclusion chromatography (Sephadex G-200 or Sepharose 4B), intrinsic fluorescence emission of α-LA bound to SUV, and scanning microcalorimetry [69][70][71][72]. It was shown that α-LA also interacts with membranes containing negatively charged lipids such as phosphatidylglycerol. Interactions of several conformers of α-LA in aqueous solution with negatively charged large unilamellar vesicles (lecithin, 1,2-dioleoylphosphatidylglycerol, dipalmitoylphosphatidyl-glycerol,) were studied by CD, infrared spectroscopy, differential scanning calorimetry, and by content leakage experiments [73][74][75].
The interaction with membranes changes physical properties of both α-LA and membranes.It was concluded that a limited expansion of conformation of α-LA occurs upon membrane association at neutral or slightly acidic pH at the physiological temperature, with a concomitant increase in tryptophan exposure to solvent and external quenchers. The conformations of the membrane-bound protein range from native-like to molten globule-like. At a low pH, α-LA penetrates the interior of the negatively charged membranes and exhibits a molten globule conformation. The degree of unsaturation of the acyl chains and the proportion of charged phospholipid species in the membranes made of neutral and acidic glycerophospholipids are determinants for the association of α-LA with liposomes and for the impermeability of the bilayer [76]. Tighter packing prevented interaction with α-LA, while unsaturation leading to looser packing promoted interaction with α-LA and leakage of contents.
7. Bactericidal, Antiviral and other Biological Activities of α-Lactalbumin and Its Fragments
α-LA and some of its fragments possess bactericidal and antiviral activities. Proteolytic digestion of α-LA by trypsin and chymotrypsin yields three peptides with bactericidal properties: LDT1 and LDT2 (tryptic digestion), and LDC (limited proteolysis with chymotrypsin) [77]. LDT2 and LDC fragments are composed of two peptide chains held together by disulfide bridges. Curiously, the bactericidal activity of these fragments depends on the presence of the disulfide bridges, and the single chain peptides derived from LDT2 or LDC by the reduction of disulfides are devoid of any bactericidal activity . The polypeptides are mostly active against Gram-positive bacteria, suggesting a possible antimicrobial function of α-LA after its partial digestion by endopeptidases.
It was found that α-lactorphin, tetrapeptide Tyr-Gly-Leu-Phe, fragment 50–53 of α-LA, obtained by enzymatic hydrolysis of α-LA by pepsin and trypsin, lowers blood pressure in adult spontaneously hypertensive rats [78]. A 35 amino acid residues long peptide, a cleaved by endopeptidase Lys C product from human α-LA (residues 59–93), induces the growth of human fetal lung fibroblast cells [79]. Two highly potent α-LA peptides with an angiotensin-converting enzyme inhibitory effect are found [80]. They correspond to the α-LA fragments 16–26 (sequence KGYGGVSLPEW) and 97–104 (DKVGINYW). Their IC50 values are as low as 0.80 and 25.2 g/mL, respectively. An α-LA folding variant with bactericidal activity against antibiotic-resistant and antibiotic-susceptible strains of Streptococcus pneumonia has been found [81].
α-LA exhibits anti-Helicobacter activities in vivo with well-demonstrated antiulcer potential in rats [82][83][84]. Esterified (methylated and ethylated) forms of α-LA are able to protect Lactococcus lactis against infection by lactococcal bacteriophages (bIL66, bIL67, and bIL170) [232] and also possess strong antiviral activity against bacteriophage M13 [85].
Yarramala et al. replaced the natural calcium ion in the binding site of bovine α-LA with lanthanum ion La3+ [86]. Surprisingly, the complex La3+-α-LA exhibited much stronger anticancer activity against breast cancer cells as compared to the BAMLET (bovine α-LA made lethal to tumor cells; see below) complex. La3+-α-LA is preferentially more toxic to MCF-7 cells as compared to KB (oral cancer) and HeLa (cervical) cells, while almost non-toxic to the healthy cells.
8. Anti-Tumorigenic Activities of α-Lactalbumin
Hakansson et al. [9][87] and Svensson et al. [88] discovered that some multimeric human α-LA derivatives served as a potent apoptosis-inducing agent with broad, yet selective, cytotoxic activity, killing all transformed, embryonic, and lymphoid cells tested. The multimeric α-LA complexed with lipids (which was called MAL) induced a loss of the mitochondrial membrane potential, mitochondrial swelling, and the release of cytochrome c, followed by activation of the caspase cascade [89]. MAL was shown to cross the plasma membrane and cytosol and enter the cell nucleus, where it induced DNA fragmentation through a direct effect at the nuclear level [90].
Similar results were obtained with HAMLET (human α-lactalbumin made lethal to tumor cells), which is a native human α-LA converted in vitro to the apoptosis-inducing folding variant by complexing the protein with unsaturated C18 fatty acids in the cis-conformation (oleic acid) [91][92][93]. The protein molecules in HAMLET are in a molten globule-like conformation. Besides human α-LA, similar anticancer activities demonstrate the oleic acid-bound forms of α-LAs from other species, such as cow, camel, and goat (BAMLET, CAMLET, and GAMLET for bovine, camel, and goat α-LA made lethal to tumor cells, respectively).
The formation of the complex α-LA-oleic acid is strongly dependent on calcium, ionic strength, and temperature [94]. The spectrofluorimetrically estimated number of oleic acid molecules irreversibly bound per α-LA molecule (after dialysis of the oleic acid-loaded preparation against water followed by lyophilization) was dependent on temperature, being equal to 2.9 at 17°C (native apo-α-LA; resulting complex referred to as LA-OA-17 state) and 9 at 45 °C (thermally unfolded apo-α-LA; LA-OA-45).
HAMLET was found to bind histones H3, H4, and H2B [95][96]. In tumor cells, HAMLET co-localizes with histones and perturbs the chromatin structure; HAMLET associates with chromatin in an insoluble nuclear fraction resistant to salt extraction. In vitro, HAMLET binds strongly to histones and impairs their deposition on DNA. It was suggested that the interaction of HAMLET with histones and chromatin in tumor cell nuclei locks the cells into the death pathway by irreversibly disrupting chromatin organization. α-LA internalizes into the cells and enters even the nucleus only when it is complexed with oleic acid [97]. It is of interest that monomeric α-LA in the absence of fatty acids is also able to bind efficiently to the primary target of HAMLET, histone H3, regardless of the Ca2+ content [98]. Thus, the modification of α-LA by oleic acid is not required for the binding to histones. The interaction of negatively charged α-LA with the basic histone stabilizes apo-α-LA and destabilizes the Ca2+-bound protein due to compensation for the excess negative charge of α-LA’s Ca2+-binding loop by positively charged residues of the histone. Co-incubation of positively charged poly-amino acids (poly-Lys and poly-Arg), used as models of histones, with α-LA resulted in effects similar to those caused by histone H3, confirming the electrostatic nature of the α-LA-histone interaction. The conformational state of α-LA in a complex with poly-Lys(Arg), named α-lactalbumin modified by poly-amino acid (LAMPA), differs from all other α-LA states characterized to date, representing an apo-like (molten globule-like) state with substantially decreased affinity for calcium ion [99].
Zherelova et al. studied interactions of the complexes of α-LA-oleic acid with small unilamellar dipalmitoylphosphatidylcholine (DPPC) vesicles and electro-excitable plasma membrane of internodal native and perfused cells of the green alga Chara corallina [100]. It was shown that oleic acid binding increases the affinity of α-LA to DPPC vesicles. Calcium association decreases protein affinity to the vesicles. The voltage clamp technique studies showed that LA-OA-17, HAMLET, and their constituents produced different modifying effects on the plasmalemmal ionic channels of the Chara corallina cells [100]. The irreversible binding of the oleic acid complexes to the plasmalemma was accompanied by changes in the activation–inactivation kinetics of developing integral transmembrane currents, suppression of the Ca2+ current and Ca2+-activated Cl− current, and by the increase in the non-specific K+ leakage currents. The latter reflected in development of nonselective permeability of the plasma membrane. The HAMLET-induced effects on the plasmalemmal currents were less pronounced and potentiated by LA-OA-17.
The treatment of tumor cells by HAMLET results in their apoptotic changes, such as caspase activation, phosphatidyl serine externalization, and chromatin condensation, but caspase inhibition or Bcl-2 over-expression not prolongs cell survival and the caspase response is Bcl-2 independent [101]. HAMLET translocates to the nuclei and binds directly to chromatin, but the death response is not related to the p53 status of the tumor cells. It was concluded that tumor cell death in response to the HAMLET treatment is independent of caspases, p53, and Bcl-2 even though HAMLET is able to activate an apoptotic response. It was revealed that HAMLET-like complexes kill some microorganisms. For example, Hakansson et al. showed that HAMLET-treated bacteria undergo cell death with mechanistic and morphologic similarities to apoptotic death of tumor cells [102].
To assess the protein’s contribution to cytotoxicity of HAMLET, Permyakov et al. prepared complexes of oleic acid with proteins, structurally and functionally distinct from α-LA [103]. Similarly to HAMLET, the resulting samples of bovine β-lactoglobulin (β-LG) and pike parvalbumin (pPA) (bLG-OA-45 and pPA-OA-45) induced Streptococcus pneumoniae D39 cell death. The activation mechanisms of Streptococcus pneumoniae death for these complexes were analogous to those for HAMLET. Cytotoxicity of the complexes against Streptococcus pneumoniae increased with oleic acid content in the sample. The IC50 value for HEp-2 cells linearly decreased with the rise in oleic acid content in the sample. Furthermore, the dependencies of HEp-2 cells viability upon oleic acid concentration in the sample for the complexes were close to that for oleic acid alone. Hence, cytotoxic action of these complexes against HEp-2 cells is induced mostly by oleic acid. Overall, the proteinaceous component of HAMLET-like complexes is not a prerequisite for their activity; the cytotoxicity of these complexes is mostly due to the action of oleic acid. Many other authors also concluded that the protein portion of the HAMLET-like complexes is not the origin of their cytotoxicity but plays a role of a delivery carrier of cytotoxic fatty acid molecules into tumor cells across the cell membrane [104][105][106][107][108].
Ho et al. presented the low-resolution solution structure of the complex of HAMLET, derived from small angle X-ray scattering (SAXS) data [109]. HAMLET shows a two-domain structure with a large globular domain and an extended part of about 2.22 in length and 1.29 nm width. The major part of α-LA accommodates well in the shape of HAMLET. However, the C-terminal residues 105 to 123 of human α-LA do not fit well into the HAMLET structure. It results in an extended conformation of HAMLET, proposed to be required to form the tumoricidal active HAMLET complex with oleic acid.
A comprehensive analysis of a series of twelve cytotoxic protein–oleic acid complexes, formed from seven structurally unrelated proteins (bovine β-lactoglobulin, pike parvalbumin, bovine serum albumin, immunoglobulin G, ovalbumin, equine lysozyme, and bovine α-lactalbumin) and prepared by different procedures, revealed that these complexes possessed the common core-shell structure, where an oily core is made of a micellar oleic acid, whereas a proteinaceous shell, which stabilizes the oleic acid micelle, is formed from the flexible, partially unfolded proteins [110]. Therefore, these liprotides (lipids and partially denatured proteins), which are potential novel anti-tumorous drugs, can be considered as molten globular containers filled with the toxic oil. Liprotides are known to increase the fluidity of membranes and transfer oleic acid to vesicles. Frislev et al. studied the effect of liprotides on the cell membrane and revealed their cytotoxicity against MCF7 cells [111]. Studies of liprotides are important because of their anti-carcinogenic properties, which are promising for their applications in the clinic [112]. In addition, liprotides can be used for drug delivery due to the ability of the micelle core to solubilize and stabilize hydrophobic compounds.
This entry is adapted from the peer-reviewed paper 10.3390/biom10091210
References
- Jackson, J.; Janszen, D.; Lönnerdal, B.; Lien, E.; Pramuk, K.; Kuhlman, C. A multinational study of α-lactalbumin concentrations in human milk. J. Nutr. Biochem. 2004, 15, 517–521.
- Renner, E. Milk and dairy products in human nutrition. In Milk and Dairy Products in Human Nutrition; John Wiley & Sons: Hoboken, NJ, USA, 1983.
- Montagne, P.; Cuilliere, M.L.; Mole, C.; Bene, M.C.; Faure, G. Immunological and nutritional composition of human milk in relation to prematurity and mother’s parity during the first 2 weeks of lactation. J. Pediatric Gastroenterol. Nutr. 1999, 29, 75–80.
- Hill, R.L.; Brew, K. Lactose synthetase. Adv. Enzymol. Relat. Areas Mol. Biol. 1975, 43, 411–490.
- Hiraoka, Y.; Segawa, T.; Kuwajima, K.; Sugai, S.; Murai, N. α-Lactalbumin—A calcium metalloprotein. Biochem. Biophys. Res. Commun. 1980, 95, 1098–1104.
- Permyakov, E.A.; Yarmolenko, V.V.; Kalinichenko, L.P.; Morozova, L.A.; Burstein, E.A. Calcium binding to α-lactalbumin: Structural rearrangement and association constant evaluation by means of intrinsic protein fluorescence changes. Biochem. Biophys. Res. Commun. 1981, 100, 191–197.
- Dolgikh, D.A.; Gilmanshin, R.I.; Brazhnikov, E.V.; Bychkova, V.E.; Semisotnov, G.V.; Venyaminov, S.Y.; Ptitsyn, O.B. α-Lactalbumin—Compact state with fluctuating tertiary structure. FEBS Lett. 1981, 136, 311–315.
- Kuwajima, K. The molten globule state of α-lactalbumin. FASEB J. 1996, 10, 102–109.
- Hakansson, A.; Zhivotovsky, B.; Orrenius, S.; Sabharwal, H.; Svanborg, C. Apoptosis Induced by a human-milk protein. Proc. Natl. Acad. Sci. USA 1995, 92, 8064–8068.
- Pellegrini, A.; Thomas, U.; Bramaz, N.; Hunziker, P.; von Fellenberg, R. Isolation and identification of three bactericidal domains in the bovine α-lactalbumin molecule. Biochim. Biophys. Acta 1999, 1426, 439–448.
- Permyakov, E.A.; Berliner, L.J. α-Lactalbumin: Structure and function. FEBS Lett. 2000, 473, 269–274.
- Permyakov, E.A. α-Lactalbumin; Nova Science Publishers: New York, NY, USA, 2005.
- Permyakov, E.A. Metalloproteomics; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2009.
- Permyakov, E.A.; Kretsinger, R.H. Calcium Binding Proteins; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2011.
- Permyakov, E.A.; Permyakov, S.E.; Breydo, L.; Redwan, E.M.; Almehdar, H.A.; Uversky, V.N. Disorder in milk proteins: α-lactalbumin. Part A. Structural properties and conformational behavior. Curr. Protein Pept. Sci. 2016, 17, 352–367.
- Permyakov, E.A.; Permyakov, S.E.; Breydo, L.; Redwan, E.M.; Almehdar, H.A.; Uversky, V.N. Disorder in milk proteins: α-lactalbumin. Part C. Pecularities of metal binding. Curr. Protein Pept. Sci. 2016, 17, 735–745.
- Uversky, V.N.; Permyakov, S.E.; Breydo, L.; Redwan, E.M.; Almehdar, H.A.; Permyakov, E.A. Disorder in milk proteins: α-lactalbumin. Part B. A multifunctional whey protein acting as an oligomeric molten globule “oil container” in the anti-tumorogenic drugs, liprotides. Curr. Protein Pept. Sci. 2016, 17, 612–628.
- Dautigny, A.; Prager, E.M.; Phamdinh, D.; Jolles, J.; Pakdel, F.; Grinde, B.; Jolles, P. cDNA and amino-acid-sequences of rainbow-trout (Oncorhynchus-Mykiss) lysozymes and their implications for the evolution of lysozyme and lactalbumin. J. Mol. Evol. 1991, 32, 187–198.
- Grobler, J.A.; Rao, K.R.; Pervaiz, S.; Brew, K. Sequences of two highly divergent canine type c lysozymes: Implications for the evolutionary origins of the lysozyme/α-lactalbumin superfamily. Arch. Biochem. Biophys. 1994, 313, 360–366.
- Nitta, K.; Sugai, S. The Evolution of lysozyme and α-lactalbumin. Eur. J. Biochem. 1989, 182, 111–118.
- Prager, E.M.; Wilson, A.C. Ancient origin of lactalbumin from lysozyme—Analysis of DNA and amino acid sequences. J. Mol. Evol. 1988, 27, 326–335.
- Threadgill, D.W.; Womack, J.E. Genomic analysis of the major bovine-milk protein genes. Nucleic Acids Res. 1990, 18, 6935–6942.
- Gallagher, D.S.; Threadgill, D.W.; Ryan, A.M.; Womack, J.E.; Irwin, D.M. Physical mapping of the lysozyme gene family in cattle. Mamm. Genome 1993, 4, 368–373.
- Davies, M.S.; West, L.F.; Davis, M.B.; Povey, S.; Craig, R.K. The gene for human α-lactalbumin is assigned to chromosome-12q13. Ann. Hum. Genet. 1987, 51, 183–188.
- Xue, Y.M.; Liu, J.N.; Sun, Z.Y.; Ma, Z.; Wu, C.L.; Zhu, D.X. α-Lactalbumin mutant acting as lysozyme. Proteins 2001, 42, 17–22.
- Chang, J.Y.; Li, L. Pathway of oxidative folding of α-lactalbumin: A model for illustrating the diversity of disulfide folding pathways. Biochemistry 2002, 41, 8405–8413.
- Barman, T.E. Purification and properties of bovine milk glycol-α-lactalbumin. Biochim. Biophys. Acta 1970, 214, 242–244.
- Brown, R.C.; Fish, W.W.; Hudson, B.G.; Ebner, K.E. Isolation and characterization of rat α-lactalbumin—Glycoprotein. Biochim. Biophys. Acta 1977, 491, 82–92.
- Pike, A.C.W.; Brew, K.; Acharya, K.R. Crystal structures of guinea-pig, goat and bovine α-lactalbumin highlight the enhanced conformational flexibility of regions that are significant for its action in lactose synthase. Structure 1996, 4, 691–703.
- Horii, K.; Saito, M.; Yoda, T.; Tsumoto, K.; Matsushima, M.; Kuwajima, K.; Kumagai, I. Contribution of Thr29 to the thermodynamic stability of goat α-lactalbumin as determined by experimental and theoretical approaches. Proteins 2001, 45, 16–29.
- Calderone, V.; Giuffrida, M.G.; Viterbo, D.; Napolitano, L.; Fortunato, D.; Conti, A.; Acharya, K.R. Amino acid sequence and crystal structure of buffalo α-lactalbumin. FEBS Lett. 1996, 394, 91–95.
- Acharya, K.R.; Ren, J.S.; Stuart, D.I.; Phillips, D.C.; Fenna, R.E. Crystal structure of human α-lactalbumin at 1.7 Å resolution. J. Mol. Biol. 1991, 221, 571–581.
- Acharya, K.R.; Stuart, D.I.; Walker, N.P.; Lewis, M.; Phillips, D.C. Refined structure of baboon α-lactalbumin at 1.7 Å resolution. Comparison with C-type lysozyme. J. Mol. Biol. 1989, 208, 99–127.
- Dolgikh, D.A.; Abaturov, L.V.; Bolotina, I.A.; Brazhnikov, E.V.; Bychkova, V.E.; Gilmanshin, R.I.; Lebedev, Y.O.; Semisotnov, G.V.; Tiktopulo, E.I.; Ptitsyn, O.B. Compact state of a protein molecule with pronounced small-scale mobility—Bovine α-lactalbumin. Eur. Biophys. J. 1985, 13, 109–121.
- Kataoka, M.; Kuwajima, K.; Tokunaga, F.; Goto, Y. Structural characterization of the molten globule of α-lactalbumin by solution X-ray scattering. Protein Sci. 1997, 6, 422–430.
- Polverino de Laureto, P.; Frare, E.; Gottardo, R.; Fontana, A. Molten globule of bovine α-lactalbumin at neutral pH induced by heat, trifluoroethanol, and oleic acid: A comparative analysis by circular dichroism spectroscopy and limited proteolysis. Proteins 2002, 49, 385–397.
- Permyakov, E.A.; Morozova, L.A.; Burstein, E.A. Cation binding effects on the pH, thermal and urea denaturation transitions in α-lactalbumin. Biophys. Chem. 1985, 21, 21–31.
- Rosner, H.I.; Redfield, C. The human α-lactalbumin molten globule: Comparison of structural preferences at pH 2 and pH 7. J. Mol. Biol. 2009, 394, 351–362.
- Bai, P.; Luo, L.; Peng, Z.Y. Side chain accessibility and dynamics in the molten globule state of α-lactalbumin: A F-19-NMR study. Biochemistry 2000, 39, 372–380.
- Redfield, C.; Schulman, B.A.; Milhollen, M.A.; Kim, P.S.; Dobson, C.M. α-Lactalbumin forms a compact molten globule in the absence of disulfide bonds. Nat. Struct. Biol. 1999, 6, 948–952.
- Permyakov, S.E.; Permyakov, E.A.; The use of the free metal—Temperature ‘phase diagrams’ for studies of single site metal binding proteins. Protein J 2007, 26, 1-12.
- Permyakov, S.E.; Khokhlova, T.I.; Uversky, V.N.; Permyakov, E.A. Analysis of Ca2+/Mg2+ selectivity in α-lactalbumin and Ca2+-binding lysozyme reveals a distinct Mg2+-specific site in lysozyme. Proteins 2010, 78, 2609–2624.
- Permyakov, A.; Ostrovsky, A.V.; Kalinichenko, L.P. Stopped flow kinetic studies of Ca(II) and Mg(II) dissociation in cod parvalbumin and bovine α-lactalbumin. Biophys. Chem. 1987, 28, 225–233.
- Murakami, ; Berliner, L.J. A distinct zinc binding site in the α-lactalbumins regulates calcium binding. Is there a physiological role for this control? Biochemistry 1983, 22, 3370–3374.
- Permyakov, E.A.; Shnyrov, V.L.; Kalinichenko, L.P.; Kuchar, A.; Reyzer, I.L.; Berliner, L.J. Binding of Zn(Ii) ions to α-lactalbumin. J. Protein Chem. 1991, 10, 577–584.
- Ren, J.S.; Stuart, D.I.; Acharya, K.R. α-Lactalbumin possesses a distinct zinc-binding site. J. Biol. Chem. 1993, 268, 19292–19298.
- Permyakov, S.E.; Veprintsev, D.B.; Brooks, C.L.; Permyakov, E.A.; Berliner, L.J. Zinc binding in bovine α-lactalbumin: Sequence homology may not be a predictor of subtle functional features. Proteins 2000, 40, 106–111.
- Saeki, K.; Arai, M.; Yoda, T.; Nakao, M.; Kuwajima, K. Localized nature of the transition-state structure in goat α-lactalbumin folding. J. Mol. Biol. 2004, 341, 589–604.
- Arai, M.; Ito, K.; Inobe, T.; Nakao, M.; Maki, K.; Kamagata, K.; Kihara, H.; Amemiya, Y.; Kuwajima, K. Fast compaction of α-lactalbumin during folding studied by stopped-flow X-ray scattering. J. Mol. Biol. 2002, 321, 121–132.
- Bushmarina, N.A.; Blanchet, C.E.; Vernier, G.; Forge, V. Cofactor effects on the protein folding reaction: Acceleration of α-lactalbumin refolding by metal ions. Protein Sci. 2006, 15, 659–671.
- Bu, Z.M.; Cook, J.; Callaway, D.J.E. Dynamic regimes and correlated structural dynamics in native and denatured α-lactalbumin. J. Mol. Biol. 2001, 312, 865–873.
- Otzen, E.; Sehgal, P.; Westh, P. α-Lactalbumin is unfolded by all classes of surfactants but by different mechanisms. J. Colloid Interface Sci. 2009, 329, 273–283.
- Singh, ; Hassan, M.I.; Islam, A.; Ahmad, F. Cooperative unfolding of residual structure in heat denatured proteins by urea and guanidinum chloride. PLoS ONE 2015, 10, e0128740.
- Goers, J.; Permyakov, S.E.; Permyakov, E.A.; Uversky, V.N.; Fink, A.L. Conformational prerequisites for α-lactalbumin fibrillation. Biochemistry 2002, 41, 12546–12551.
- Polverino de Laureto, P.; Frare, E.; Battaglia, F.; Mossuto, M.F.; Uversky, V.N.; Fontana, A. Protein dissection enhances the amyloidogenic properties of α-lactalbumin. FEBS J. 2005, 272, 2176–2188.
- Buccarielli, S.; Sayedi, E.S.; Osella, S.; Trzaskowsli, B.; Vissing, K.J.; Vestergaard, B.; Fodera, V. Disentangling the role of solvent polarity and protein solvation in folding and self-assembly of α-lactalbumin. J. Colloid Interface Sci. 2020, 561, 749–761.
- Arroyo-Maya, I.J.; Rodiles-López, J.O.; Cornejo-Mazón, M.; Gutiérrez-López, G.F.; Hernández-Arana, A.; Toledo-Núñez, C.; Barbosa-Cánovas, G.V.; Flores-Flores, J.O.; Hernández-Sánchez, H.J. Effect of different treatments on the ability of α-lactalbumin to form nanoparticles. Dairy Sci. 2012, 95, 6204–6214.
- Zhang, B.; Luo, Y.; Wang, Q. Development of silver/α-lactalbumin nano-composites: A new approach to reduce silver toxicity. Int. J. Antimicrob. Agents 2011, 38, 502–509.
- Ipsen, R.; Otte, J.; Qvist, K.B. Molecular self-assembly of partially hydrolysed α-lactalbumin resulting in strong gels with a novel microstructure. J. Dairy Res. 2001, 68, 277–286.
- Geng, X.L.; Kain Kirkensgaard, J.J.; Arleth, L.; Otte, J.; Ipsen. R. The influence of pH, protein concentration and calcium ratio on the formation and structure of nanotubes from partially hydrolyzed bovine α-lactalbumin. Soft Matter 2019, 15, 4787–4796.
- Permyakov, E.A.; Kreimer, D.I. Effects of pH, Temperature and Ca2+ content on the conformation of α-lactalbumin in a medium modeling physiological conditions. Gen. Physiol. Biophys. 1986, 5, 377–390.
- Vanderheeren, G.; Hanssens, I.; Noyelle, K.; Van Dael, H.; Joniau, M. The perturbations of the native state of goat α-lactalbumin induced by 1,1’-bis(4-anilino-5-naphthalenesulfonate) are Ca2+-dependent. Biophys. J. 1998, 75, 2195–2204.
- Singh, S.K.; Kishore, N. Elucidating the binding thermodynamics of 8-anilino-1-naphthalene sulfonic acid with the A-state of α-lactalbumin: An isothermal titration calorimetric investigation. Biopolymers 2006, 83, 205–212.
- Permyakov, E.A.; Grishchenko, V.M.; Kalinichenko, L.P.; Orlov, N.Y.; Kuwajima, K.; Sugai, S. Calcium-regulated interactions of human α-lactalbumin with bee venom melittin. Biophys. Chem. 1991, 39, 111–117.
- Cawthern, K.M.; Permyakov, E.; Berliner, L.J. Membrane-bound states of α-lactalbumin: Implications for protein stability and conformation. Protein Sci. 1996, 5, 1394–1405.
- Cawthern, K.M.; Narayan, M.; Chaudhuri, D.; Permyakov, E.A.; Berliner, L.J. Interactions of α-lactalbumin with fatty acids and spin label analogs. J. Biol. Chem. 1997, 272, 30812–30816.
- Nigen, M.; Croguennec, T.; Renard, D.; Bouhallab, S. Temperature affects the supramolecular structures resulting from α-lactalbumin-lysozyme interaction. Biochemistry 2007, 46, 1248–1255.
- Nigen, M.; Gaillard, C.; Croguennec, T.; Madec, N.; Bouhallab, S. Dynamic and supramolecular organisation of α-lactalbumin/lysozyme microspheres: A microscopic study. Biophys. Chem. 2010, 146, 30–35.
- Berliner, L.J.; Koga, K. α-Lactalbumin binding to membranes—Evidence for a partially buried protein. Biochemistry 1987, 26, 3006–3009.
- Permyakov, A.; Kreimer, D.I.; Kalinichenko, L.P.; Shnyrov, V.L. Effects of calcium and magnesium-ions on the interactions of calcium-binding proteins parvalbumin and α-lactalbumin with dipalmitoylphosphatidylcholine vesicles. Biofizika 1988, 33, 465–470.
- Permyakov, A.; Kreimer, D.I.; Kalinichenko, L.P.; Shnyrov, V.L. Interactions of calcium-binding proteins, parvalbumin and α-lactalbumin, with dipalmitoyl phosphatidyl choline vesicles. Gen. Physiol. Biophys. 1988, 7, 95–107.
- Grishchenko, M.; Kalinichenko, L.P.; Deikus, G.Y.; Veprintsev, D.B.; Cawthern, K.M.; Berliner, L.J.; Permyakov, E.A. Interactions of α-lactalbumins with lipid vesicles studied by tryptophan fluorescence. Biochem. Mol. Biol. Int. 1996, 38, 453–466.
- Bañuelos, S.; Muga, A. Binding of molten globule-like conformations to lipid bilayers—Structure of native and partially folded α-lactalbumin bound to model membranes. J. Biol. Chem. 1995, 270, 29910–29915.
- Bañuelos, ; Muga, A. Structural requirements for the association of native and partially folded conformations of α-lactalbumin with model membranes. Biochemistry 1996, 35, 3892–3898.
- Bañuelos, S.; Muga, A. Interaction of native and partially folded conformations of α-lactalbumin with lipid bilayers: Characterization of two membrane-bound states. FEBS Lett. 1996, 386, 21–25.
- Rødland, I.; Halskau, Ø.; Martínez, ; Holmsen, H. α-Lactalbumin binding and membrane integrity—Effect of charge and degree of unsaturation of glycerophospholipids. Biochim. Biophys. Acta 2005, 1717, 11–20.
- Pellegrini, Antimicrobial peptides from food proteins. Curr. Pharm. Des. 2003, 9, 1225–1238.
- Sipola, M.; Finckenberg, P.; Vapaatalo, H.; Pihlanto-Leppala, A.; Korhonen, H.; Korpela, R.; Nurminen, M.L. α-Lactorphin and β-lactorphin improve arterial function in spontaneously hypertensive rats. Life Sci. 2002, 71, 1245–1253.
- Kanda, Y.; Hisayasu, S.; Abe, Y.; Katsura, K.; Mashimo, K. Growth-active peptides are produced from α-lactalbumin and lysozyme. Life Sci. 2007, 81, 449–457.
- Tavares, T.; Contreras, M.D.; Amorim, M.; Pintado, M.; Recio, I.; Malcata, F.X. Novel whey-derived peptides with inhibitory effect against angiotensin-converting enzyme: In vitro effect and stability to gastrointestinal enzymes. Peptides 2011, 32, 1013–1019.
- Hakansson, A.; Svensson, M.; Mossberg, A.K.; Sabharwal, H.; Linse, S.; Lazou, I.; Lonnerdal, S.; Svanborg, C. A folding variant of α-lactalbumin with bactericidal activity against Streptococcus pneumoniae. Mol. Microbiol. 2000, 35, 589–600.
- Matsumoto, H.; Shimokawa, Y.; Ushida, Y.; Toida, T.; Hayasawa, H. New biological function of bovine α-lactalbumin: Protective effect against ethanol- and stress-induced gastric mucosal injury in rats. Biosci. Biotechnol. Biochem. 2001, 65, 1104–1111.
- Mezzaroba, L.F.H.; Carvalho, J.E.; Ponezi, A.N.; Antônio, M.A.; Monteiro, K.M.; Possenti, A.; Sgarbieri, V.C. Antiulcerative properties of bovine α-lactalbumin. Int. Dairy J. 2006, 16, 1005–1012.
- Sachdeva, A; Rawat, S.; Nagpal, J. Efficacy of fermented milk and whey proteins in Helicobacter pylori eradication: A review. World J. Gastroenterol. 2014, 20, 724–737.
- Sitohy, M.; Chobert, J.M.; Karwowska, U.; Gozdzicka-Jozefiak, A.; Haertle, T. Inhibition of bacteriophage m13 replication with esterified milk proteins. J. Agric. Food Chem. 2006, 54, 3800–3806.
- Yarramala, D.S.; Prakash, P.; Ranade, D.S.; Doshi, S.; Kulkarni, P.P.; Bhaumik, P.; Rao, C.P. Cytotoxicity of apo bovine α-lactalbumin complexed with La3+ on cancer cells supported by its high resolution crystal structure. Sci. Rep. 2019, 9, 1780.
- Hakansson, A.; Andreasson, J.; Zhivotovsky, B.; Karpman, D.; Orrenius, S.; Svanborg, C. Multimeric α-lactalbumin from human milk induces apoptosis through a direct effect on cell nuclei. Exp. Cell Res. 1999, 246, 451–460.
- Svensson, M.; Sabharwal, H.; Hakansson, A.; Mossberg, A.K.; Lipniunas, P.; Leffler, H.; Svanborg, C.; Linse, S. Molecular characterization of α-lactalbumin folding variants that induce apoptosis in tumor cells. J. Biol. Chem. 1999, 274, 6388–6396.
- Kohler, C.; Hakansson, A.; Svanborg, C.; Orrenius, S.; Zhivotovsky, B. Protease activation in apoptosis induced by MAL. Exp. Cell Res. 1999, 249, 260–268.
- Svensson, M.; Hakansson, A.; Mossberg, A.K.; Linse, S.; Svanborg, C. Conversion of α-lactalbumin to a protein inducing apoptosis. Proc. Natl. Acad. Sci. USA 2000, 97, 4221–4226.
- Svanborg, C.; Agerstam, H.; Aronson, A.; Bjerkvig, R.; Duringer, C.; Fischer, W.; Gustafsson, L.; Hallgren, O.; Leijonhuvud, I.; Linse, S.; et al. HAMLET kills tumor cells by an apoptosis-like mechanism—Cellular, molecular, and therapeutic aspects. Adv. Cancer Res. 2003, 88, 1–29.
- Gustafsson, L; Hallgren, O.; Mossberg, A.K.; Pettersson, J.; Fischer, W.; Aronsson, A.; Svanborg, C. HAMLET kills tumor cells by apoptosis: Structure, cellular mechanisms, and therapy. J. Nutr. 2005, 135, 1299–1303.
- Svensson, M.; Mossberg, A.K.; Pettersson, J.; Linse, S.; Svanborg, C. Lipids as cofactors in protein folding: Stereo-specific lipidprotein interactions are required to form HAMLET human α-lactalbumin made lethal to tumor cells. Protein Sci. 2003, 12, 2805–2814.
- Knyazeva, E.L.; Grishchenko, V.M.; Fadeev, R.S.; Akatov, V.S.; Permyakov, S.E.; Permyakov, E.A. Who is Mr. HAMLET? Interaction of human α-lactalbumin with monomeric oleic acid. Biochemistry 2008, 47, 13127–13137.
- Duringer, C; Hamiche, A.; Gustafsson, L.; Kimura, H.; Svanborg, C. HAMLET interacts with histones and chromatin in tumor cell nuclei. J. Biol. Chem. 2003, 278, 42131–42135.
- Brest, P; Gustafsson, M.; Mossberg, A.K.; Gustafsson, L.; Duringer, C.; Hamiche, A.; Svanborg, C. Histone deacetylase inhibitors promote the tumoricidal effect of HAMLET. Cancer Res. 2007, 67, 11327–11334.
- Mercer, N.; Ramakrishnan, B.; Boeggeman, E.; Qasba, P.K. Applications of site-specific labeling to study HAMLET, a tumoricidal complex of α-lactalbumin and oleic acid. PLoS ONE 2011, 6, e26093.
- Permyakov, E.; Pershikova, I.V.; Khokhlova, T.I.; Uversky, V.N.; Permyakov, E.A. No need to be HAMLET or BAMLET to interact with histories: Binding of monomeric α-lactalbumin to histories and basic poly-amino acids. Biochemistry 2004, 43, 5575–5582.
- Permyakov, S.E.; Pershikova, I.V.; Zhadan, A.P.; Goers, J.; Bakunts, A.G.; Uversky, V.N.; Berliner, L.J.; Permyakov, E.A. Conversion of human α-lactalbumin to an apo-like state in the complexes with basic poly-amino acids: Toward understanding of the molecular mechanism of antitumor action of HAMLET. J. Proteome Res. 2005, 4, 564–569.
- Zherelova, O.M.; Kataev, A.A.; Grishchenko, V.M.; Knyazeva, E.L.; Permyakov, S.E.; Permyakov, E.A. Interaction of antitumor α-lactalbumin-oleic acid complexes with artificial and natural membranes. J. Bioenerg. Biomembr. 2009, 41, 229–237.
- Hallgren, O.; Gustafsson, L.; Irjala, H.; Selivanova, G.; Orrenius, S.; Svanborg, C. HAMLET triggers apoptosis but tumor cell death is independent of caspases, Bcl-2 and p53. Apoptosis 2006, 11, 221–233.
- Hakansson, P.; Roche-Hakansson, H.; Mossberg, A.K.; Svanborg, C. Apoptosis-like death in bacteria induced by HAMLET, a human milk lipid-protein complex. PLoS ONE 2011, 6, e17717.
- Permyakov, S.E.; Knyazeva, E.L.; Khasanova, L.M.; Fadeev, R.S.; Zhadan, A.P.; Roche-Hakansson, H.; Hakansson, A.P.; Akatov, V.S.; Permyakov, E.A. Oleic acid is a key cytotoxic component of HAMLET-like complexes. Biol. Chem. 2012, 393, 85–92.
- Nakamura, T.; Aizawa, T.; Kariya, R.; Okada, S.; Demura, M.; Kawano, K.; Makabe, K.; Kuwajima, K. Molecular mechanisms of the cytotoxicity of human α-lactalbumin made lethal to tumor cells (HAMLET) and other protein-oleic acid complexes. J. Biol. Chem. 2013, 288, 14408–14416.
- Fontana, A.; Spolaore, B.; Polverino de Laureto, P. The biological activities of protein/oleic acid complexes reside in the fatty acid. Biochim. Biophys. Acta 2013, 1834, 1125–1143.
- Brinkmann, R.; Thiel, S.; Otzen, D.E. Protein-fatty acid complexes: Biochemistry, biophysics and function. FEBS J. 2013, 280, 1733–1749.
- Hoque, M.; Dave, S.; Gupta, P.; Saleemuddin, M. Oleic acid may be the key contributor in the BAMLET-induced erythrocyte hemolysis and tumoricidal action. PLoS ONE 2013, 8, e68390.
- Rath, E.M.; Duff, A.P.; Håkansson, A.P.; Vacher, C.S.; Liu, G.J.; Knott, R.B.; Church, W.B. Structure and potential cellular targets of HAMLET-like anti-cancer compounds made from milk components. J. Pharm. Pharm. Sci. 2015, 18, 773–824.
- Ho, C.S.; Rydstrom, A.; Manimekalai, M.S.; Svanborg, C.; Grüber, G. Low resolution solution structure of HAMLET and the importance of its α-domains in tumoricidal activity. PLoS ONE 2012, 7, e53051.
- Kaspersen, D.; Pedersen, J.N.; Hansted, J.G.; Nielsen, S.B.; Sakthivel, S.; Wilhelm, K.; Nemashkalova, E.L.; Permyakov, S.E.; Permyakov, E.A.; Pinto Oliveira, C.L.; et al. Generic structures of cytotoxic liprotides: Nano-sized complexes with oleic acid cores and shells of disordered proteins. ChemBioChem 2014, 15, 2693–2702.
- Frislev, H.S.; Boye, T.L.; Nylandsted, J.; Otzen, D. Liprotides kill cancer cells by disrupting the plasma membrane. Sci. Rep. 2017, 7, 15129.
- Pedersen, N.; Frislev, H.K.S.; Pedersen, J.S.; Otzen, D. Structures and mechanisms of formation of liprotides. Biochim. Biophys. Acta Proteins Proteom. 2020, 1868, 140505, doi:10.1016/ j.bbapap.2020.140505.
