4. Relationships between Iceland and NWT Samples
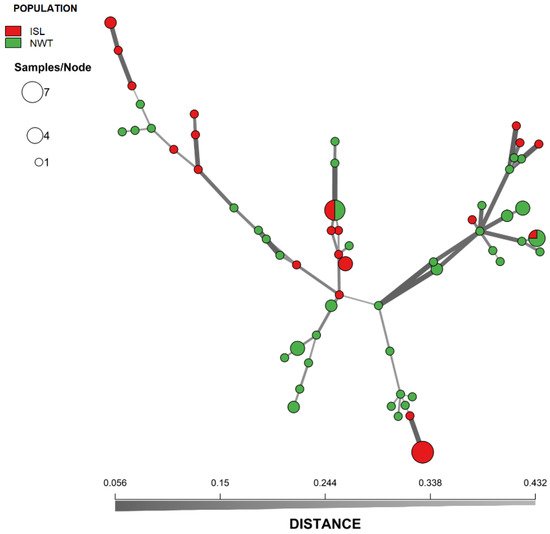
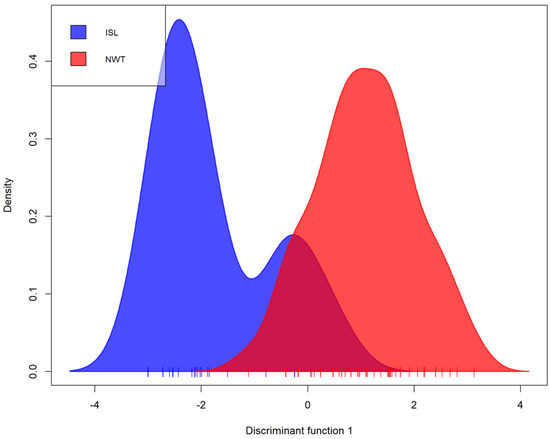
It was analyzed the relationships among A. fumigatus isolates from Iceland and NWT. Bruvo’s genetic distance was calculated between isolates of both populations and visualized through an MSN (Figure 1). Overall, while it was observed some geographic clustering among MLGs, isolates from the two arctic populations were intermixed. For example, two different MLGs were shared between Iceland and NWT populations. One of these two MLGs was shared among six strains with three isolates from each of the two regional populations. The other was shared among four isolates (one from Iceland and three from NWT). A DAPC of the Iceland and NWT samples yielded comparable results, which both showed some geographic clustering as well as overlaps between these two regions (Figure 2). Results from AMOVA indicated that while the majority (86.8%) of the genetic variations were found within the two regional populations, 13.2% of the total observed genetic variance was found between these two populations (ϕpt = 0.132, p = 0.001). Together, these results are consistent with an overall statistically significant genetic differentiation between these two arctic populations.
Figure 1. Minimum-spanning network showing the genetic relationship between MLGs of A. fumigatus from Iceland and Northwest Territory in Canada. The genetic distance between MLGs was calculated using Bruvo’s genetic distance from the nine microsatellite loci that incorporates the stepwise mutation model. Each node represents one or more identical MLGs, where node size corresponds to the number of strains for each MLG. Nodes that are more genetically similar have darker and thicker edges, whereas nodes genetically distant have lighter and thinner edges.
Figure 2. Discriminant analysis of principal components (DAPC) of Iceland and NWT A. fumigatus populations representing the first discriminant function in an individual density plot. Isolates were genotyped at nine microsatellite loci and clone corrected, totaling 60 multilocus genotypes. NWT—Northwest Territories, ISL—Iceland.
5. Relationship between the Arctic Populations to Additional Global A. fumigatus Populations
To determine how the arctic populations fit in the global context, the allelic and genotypic diversities of 12 other geographic populations totaling 2339 A. fumigatus MLGs were compared to the Iceland and NWT A. fumigatus populations. Within the 2423 MLGs, the number of private alleles present only within Iceland and NWT was 2 and 1 respectively. In Iceland, these two private alleles were #116 and #117 at the 3A locus. In NWT, the private allele was #57 at the 3B locus. Together, the allelic distribution results suggest limited novel genetic diversity in the arctic soil samples.
To determine the genetic relationships between the 2 arctic populations and the 12 other geographic populations, ϕpt was calculated between all pairwise population combinations (Table 3). For this analysis, the dataset was also clone-corrected to remove the influence of multiple clonal MLGs in determining genetic differentiation between populations. Overall, the analyses revealed that the two arctic populations of A. fumigatus were significantly different from most other geographic populations (Table 3). Specifically, before clonal correction, only the Belgian and French populations showed insignificant difference to the Iceland population. After clonal correction, only the Indian and French populations were not significantly differentiated from the Iceland population while the Norwegian and New Zealand populations were not significantly different from the NWT population.
Table 3. Pairwise differentiations between Iceland and Northwest Territories A. fumigatus populations to each other and 12 A. fumigatus populations from Eurasia, Oceania, and North America. NWT/ISL row represents the Iceland or Northwest Territories population being compared to the other. NWT—Northwest Territories, ISL—Iceland, CMR—Cameroon, CAN—Hamilton, Ontario, Canada, BEL—Belgium, FRA—France, DEU—Germany, IND—India, NLD—Netherlands, NOR—Norway, NZL—New Zealand, ESP—Spain, CHE—Switzerland, and USA—United States.
| Pairwise ϕpt |
| |
Iceland |
Northwest Territories |
| Country |
Original Dataset |
Clone Corrected |
Original Dataset |
Clone Corrected |
| NWT/ISL |
0.132 ** |
0.083 * |
0.132 ** |
0.083 * |
| CMR |
0.580 *** |
0.629 *** |
0.598 *** |
0.588 *** |
| CAN |
0.412 *** |
0.439 *** |
0.337 *** |
0.327 *** |
| BEL |
0.020 |
0.048 * |
0.178 *** |
0.041 ** |
| FRA |
0.028 |
0.012 |
0.070 ** |
0.050 * |
| DEU |
0.170 *** |
0.192 *** |
0.285 *** |
0.206 *** |
| IND |
0.130 *** |
0.052 |
0.213 *** |
0.056 * |
| NLD |
0.054 ** |
0.069 * |
0.088 *** |
0.062 ** |
| NOR |
0.152 *** |
0.050 * |
0.037 ** |
0.014 |
| NZL |
0.067 ** |
0.123 ** |
0.051 ** |
0.024 |
| ESP |
0.082 ** |
0.074 * |
0.122 *** |
0.069 *** |
| CHE |
0.089 ** |
0.100 ** |
0.063 *** |
0.036 * |
| USA |
0.162 *** |
0.146 ** |
0.129 *** |
0.051 ** |
* p-value = 0.05, ** p-value = 0.01, *** p-value = 0.001.
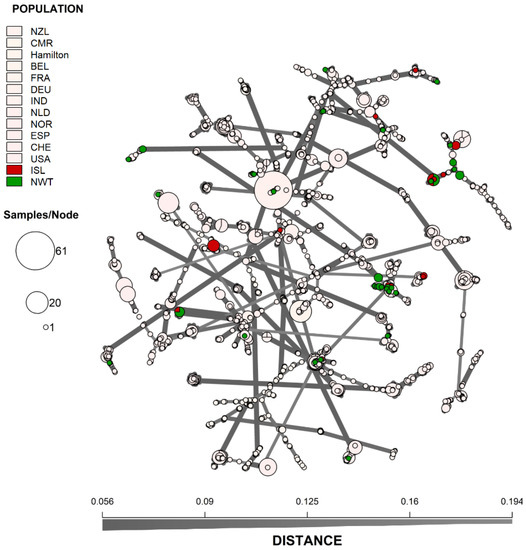
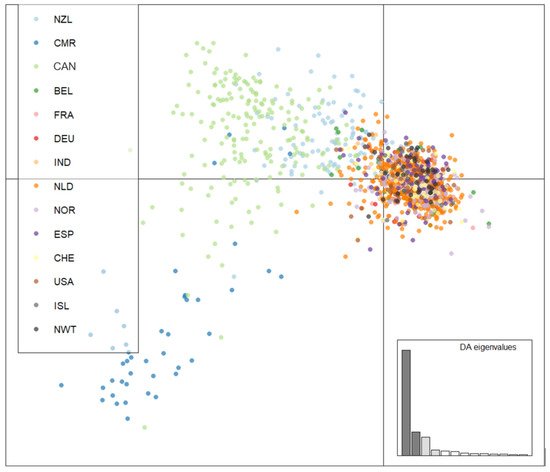
It analyzed the genotypic similarities among individual MLGs from the 14 geographic populations. An MSN of all 2416 strains was generated to visualize the genetic distance among the MLGs (Figure 3). To highlight the distribution of the arctic isolates, the 12 previously surveyed A. fumigatus populations were coloured white. Overall, while the MLGs from the arctic regions showed some geographic clustering, these arctic genotypes were broadly embedded in the global genotype network. This overall pattern was similarly supported by DAPC analyses (Figure 4) where MLGs from Iceland and NWT clustered with those from Eurasia and the United States but were different from the Hamilton, Ontario, Canada population as well as from the Cameroonian population.
Figure 3. Minimum-spanning network showing the genetic relationship between MLGs from Iceland and Northwest Territory to 12 other A. fumigatus geographic populations. The genetic distance between MLGs was calculated using Bruvo’s genetic distance based on the nine microsatellite loci that incorporates the stepwise mutation model. Each node represents one or more identical MLGs, where the node size corresponds to the number of identical MLGs. Nodes that are more genetically similar have darker and thicker edges, whereas nodes genetically distant have lighter and thinner edges. NWT—Northwest Territories, ISL—Iceland, CMR—Cameroon, CAN—Hamilton, Ontario, Canada, BEL—Belgium, FRA—France, DEU—Germany, IND—India, NLD—Netherlands, NOR—Norway, NZL—New Zealand, ESP—Spain, CHE—Switzerland, and USA—United States.
Figure 4. Genetic clustering using discriminant analysis of principal components (DAPC) of Iceland, NWT, Eurasian, North American, and Oceanian A. fumigatus populations. Isolates were genotyped at nine microsatellite loci and clone corrected, totaling 1703 unique multilocus genotypes. Populations were clustered according to population of origin. NWT—Northwest Territories, ISL—Iceland, CMR—Cameroon, CAN—Hamilton, Ontario, Canada, BEL—Belgium, FRA—France, DEU—Germany, IND—India, NLD—Netherlands, NOR—Norway, NZL—New Zealand, ESP—Spain, CHE—Switzerland, and USA—United States.
6. Susceptibility Testing
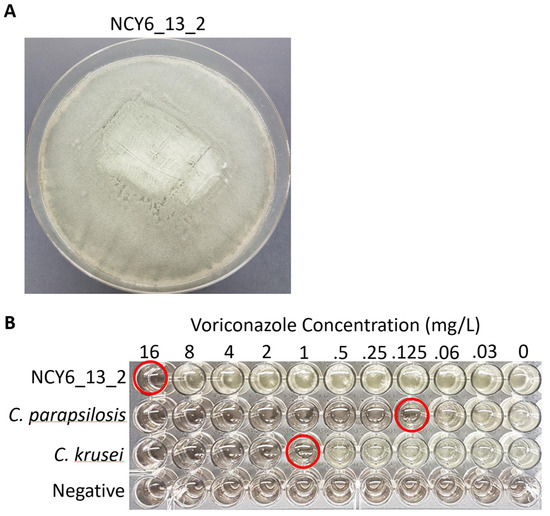
Antifungal susceptibility testing using voriconazole and itraconazole was conducted for all isolates from the two arctic populations (Table 4). One isolate resistant to both triazoles was identified within NWT but no resistant strain was identified from Iceland (Figure 5). The sequence analysis revealed that this triazole-resistant strain NCY6_13_2 had the TR34/L98H mutation within the cyp51A gene. Interestingly, this strain had a near identical genotype at the nine STR loci to a clonal population of triazole-resistant strains discovered from India in 2012, differing by only one repeat unit at the 3A locus and with identical alleles at the other eight STR loci (Table 5). Furthermore, all the A. fumigatus isolates in this Indian clone contained the identical TR34/L98H mutation within the cyp51A gene as the strain from NWT in the sample.
Figure 5. Radial growth and voriconazole susceptibility of triazole resistant A. fumigatus strain NCY6_13_2. (A) Growth morphology of A. fumigatus strain NCY6_13_2 on Malt extract agar after 5-day incubation at 35°C. (B) Voriconazole susceptibility of NCY6_13_2. Two control strains, Candida parasilosis ATCC® 22019 and C. krusei ATCC® 6258, were used to verify the susceptibility results. A negative control that was not inoculated with any microbial strain was included in the bottom row. Susceptibility endpoints are circled in red for each strain.
Table 4. Triazole antifungal susceptibility of the 32 and 52 A. fumigatus isolates from Iceland and NWT, respectively, to itraconazole and voriconazole. Numbers in the table refer to the number of strains with the respective minimum inhibitory concentrations for each of the two drugs in each geographic population.
| Country/Territory |
Triazole |
Minimum Inhibitory Concentration (mg/L) |
| 0.125 |
0.25 |
0.5 |
1 |
2 |
16 |
| Iceland |
Itraconazole |
0 |
0 |
24 |
8 |
0 |
0 |
| |
Voriconazole |
2 |
29 |
1 |
0 |
0 |
0 |
| Canada/NWT |
Itraconazole |
0 |
0 |
35 |
16 |
0 |
1 |
| |
Voriconazole |
0 |
27 |
24 |
0 |
0 |
1 |
Table 5. Short tandem repeat genotype of the A. fumigatus strain with the TR34/L98H mutation in the triazole-target gene cyp51A obtained from Northwest Territories in Canada. Alleles at eight of the nine STR loci in this strain were identical to those of the large Indian triazole-resistant clonal population.
| Strain |
Region |
2A |
2B |
2C |
3A |
3B |
3C |
4A |
4B |
4C |
| NCY6-13_2 |
NWT |
14 |
20 |
9 |
31 |
8 |
10 |
8 |
10 |
28 |
| 1042/09 |
India |
14 |
20 |
9 |
31 |
9 |
10 |
8 |
10 |
28 |