Aeromonas spp. are generally found in aquatic environments, although they have also been isolated from both fresh and processed food. These Gram-negative, rod-shaped bacteria are mostly infective to poikilothermic animals, although they are also considered opportunistic pathogens of both aquatic and terrestrial homeotherms, and some species have been associated with gastrointestinal and extraintestinal septicemic infections in humans. Several cell-surface glucans have been shown to contribute to colonization and survival of Aeromonas pathogenic strains in different hosts, playing important roles in bacterial–host interactions related to pathogenesis These include lipopolysaccharide (LPS), capsule, α-glucan, and glycosylated polar and lateral flagella.
- Aeromonas
- capsule polysaccharide
- α-glucan
- glycosylation
- O-antigen
- LPS
1. Introduction
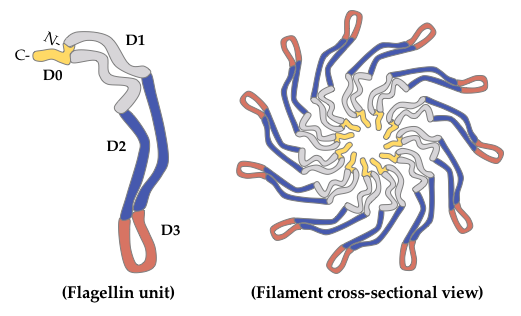
2. Glycosylated Flagella
3. Lipopolysaccharide
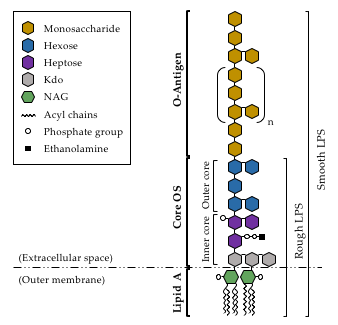
Prior to its attachment to the lipid-A-core-OS structure of LPS, before the whole complex is exported to the external side of the outer membrane, the O-antigen needs to be fully synthesized. O-antigen biosynthesis begins with the generation of the lipid-linked glycan intermediate undecaprenyl phosphate (UndP) [49] and, once generated, a sugar transfer reaction takes place. In this regard, A. piscicola AH-3 has been shown to use WecP to transfer N-acetyl-galactosamine [50]. Following this reaction, the O-antigen unit is flipped across the bacterial inner membrane by the Wzx protein [51], and assembled on the periplasmic side of the inner membrane by the Wzy O-antigen polymerase [52]. The O-antigen is elongated until it reaches the final polymer length, in a process regulated by the O-antigen chain length regulator Wzz [53]. A. piscicolaAH-3 (O:34) has been shown to follow the Wzx/Wzy-dependent pathway for O-antigen assembly, as both wzy and wzx genes are found in the O-antigen gene cluster of this strain [54]. Interestingly, although the first sugar of the O-antigen repeating unit seems to be determinant for the generation of glycosidic bonds [55], the Wzy enzyme of A. piscicola AH-3 (O:34) has been shown to be permissive with this first sugar at the non-reducing end [56].
4. Capsular Polysaccharide
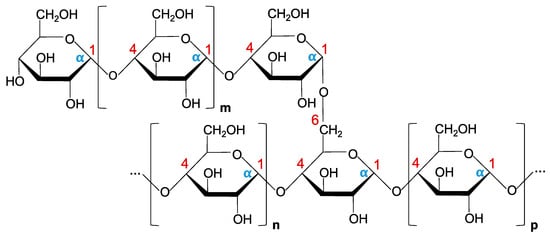
5. α-Glucan
6. Future Perspectives and Concluding Remarks
This entry is adapted from the peer-reviewed paper 10.3390/md19110649
References
- Martin-Carnahan, A.; Joseph, S.W. Order XII. Aeromonadales ord. nov. In Bergey’s Manual® of Systematic Bacteriology; Brenner, D.J., Krieg, N.R., Staley, J.T., Eds.; Springer: Boston, MA, USA, 2005; pp. 556-587.
- Janda, J.M.; Abbott, S.L.; The Genus Aeromonas : Taxonomy, Pathogenicity, and Infection. Clinical Microbiology Reviews 2010, 23, 35-73, 10.1128/cmr.00039-09.
- Majeed, K.N.; Egan, A.F.; Rae, I.C. Mac; Production of exotoxins by Aeromonas spp. at 5°C. Journal of Applied Bacteriology 1990, 69, 332-337, 10.1111/j.1365-2672.1990.tb01524.x.
- Galindo, C.L.; Chopra, A.K. Aeromonas and Plesiomonas species. In Food Microbiology: Fundamentals and Frontiers; Doyle, M.P., Beuchat, L.R., Montville, T.J., Eds.; ASM Press: Washington, DC, USA, 2007; pp. 381-400.
- Monette, S.; Dallaire, A.D.; Mingelbier, M.; Groman, D.; Uhland, C.; Richard, J.P.; Paillard, G.; Johannson, L.M.; Chivers, D.P.; Ferguson, H.W.; et al. Massive Mortality of Common Carp (Cyprinus carpio carpio) in the St. Lawrence River in 2001: Diagnostic Investigation and Experimental Induction of Lymphocytic Encephalitis. Veterinary Pathology 2006, 43, 302-310, 10.1354/vp.43-3-302.
- Rahman, M.; Colque-Navarro, P.; Kühn, I.; Huys, G.; Swings, J.; Möllby, R.; Identification and Characterization of Pathogenic Aeromonas veronii Biovar Sobria Associated with Epizootic Ulcerative Syndrome in Fish in Bangladesh. Applied and Environmental Microbiology 2002, 68, 650-655, 10.1128/aem.68.2.650-655.2002.
- Janda, J.M.; Abbott, S.L.; Evolving Concepts Regarding the Genus Aeromonas: An Expanding Panorama of Species, Disease Presentations, and Unanswered Questions. Clinical Infectious Diseases 1998, 27, 332-344, 10.1086/514652.
- Teunis, P.; Figueras, M.J.; Reassessment of the Enteropathogenicity of Mesophilic Aeromonas Species. Frontiers in Microbiology 2016, 7, 1395, 10.3389/fmicb.2016.01395.
- Pal, M.; Ayele, Y.; Durglishvili, N.; Emerging Role of Aeromonas hydrophila as a Foodborne Pathogen of Public Health Concern. EC Microbiology 2020, 16, 55-58, .
- Parker, J.L.; Shaw, J.G.; Aeromonas spp. clinical microbiology and disease. Journal of Infection 2011, 62, 109-118, 10.1016/j.jinf.2010.12.003.
- Fernández-Bravo, A.; Figueras, M.J.; An Update on the Genus Aeromonas: Taxonomy, Epidemiology, and Pathogenicity. Microorganisms 2020, 8, 129-167, 10.3390/microorganisms8010129.
- Schäffer, C.; Messner, P.; Emerging facets of prokaryotic glycosylation. FEMS Microbiology Reviews 2017, 41, 49-91, 10.1093/femsre/fuw036.
- Kirov, S.M.; Tassell, B.C.; Semmler, A.B.T.; O’Donovan, L.A.; Rabaan, A.A.; Shaw, J.G.; Lateral Flagella and Swarming Motility in Aeromonas Species. Journal of Bacteriology 2002, 184, 547-555, 10.1128/jb.184.2.547-555.2002.
- Macnab, R.M.; How Bacteria Assemble Flagella. Annual Review of Microbiology 2003, 57, 77-100, 10.1146/annurev.micro.57.030502.090832.
- Canals, R.; Ramirez, S.; Vilches, S.; Horsburgh, G.; Shaw, J.G.; Tomás, J.M.; Merino, S.; Polar Flagellum Biogenesis in Aeromonas hydrophila. Journal of Bacteriology 2006, 188, 542-555, 10.1128/jb.188.2.542-555.2006.
- Canals, R.; Altarriba, M.; Vilches, S.; Horsburgh, G.; Shaw, J.G.; Tomás, J.M.; Merino, S.; Analysis of the Lateral Flagellar Gene System of Aeromonas hydrophila AH-3. Journal of Bacteriology 2006, 188, 852-862, 10.1128/jb.188.3.852-862.2006.
- Gavín, R.; Rabaan, A.A.; Merino, S.; Tomás, J.M.; Gryllos, I.; Shaw, J.G.; Lateral flagella of Aeromonas species are essential for epithelial cell adherence and biofilm formation. Molecular Microbiology 2002, 43, 383-397, 10.1046/j.1365-2958.2002.02750.x.
- Yonekura, K.; Maki-Yonekura, S.; Namba, K.; Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature 2003, 424, 643-650, 10.1038/nature01830.
- Tabei, S.M.B.; Hitchen, P.G.; Day-Williams, M.J.; Merino, S.; Vart, R.; Pang, P.-C.; Horsburgh, G.J.; Viches, S.; Wilhelms, M.; Tomás, J.M.; et al. An Aeromonas caviae Genomic Island Is Required for both O-Antigen Lipopolysaccharide Biosynthesis and Flagellin Glycosylation. Journal of Bacteriology 2009, 191, 2851-2863, 10.1128/jb.01406-08.
- Merino, S.; Wilhelms, M.; Tomás, J.M.; Role of Aeromonas hydrophila Flagella Glycosylation in Adhesion to Hep-2 Cells, Biofilm Formation and Immune Stimulation. International Journal of Molecular Sciences 2014, 15, 21935-21946, 10.3390/ijms151221935.
- Wilhelms, M.; Fulton, K.M.; Twine, S.M.; Tomás, J.M.; Merino, S.; Differential Glycosylation of Polar and Lateral Flagellins in Aeromonas hydrophila AH-3. Journal of Biological Chemistry 2012, 287, 27851-27862, 10.1074/jbc.m112.376525.
- Parker, J.L.; Day-Williams, M.J.; Tomas, J.M.; Stafford, G.P.; Shaw, J.G.; Identification of a putative glycosyltransferase responsible for the transfer of pseudaminic acid onto the polar flagellin of Aeromonas caviae Sch3N. Microbiology Open 2012, 1, 149-160, 10.1002/mbo3.19.
- Rabaan, A.A.; Gryllos, I.; Tomás, J.M.; Shaw, J.G.; Motility and the Polar Flagellum Are Required for Aeromonas caviae Adherence to HEp-2 Cells. Infection and Immunity 2001, 69, 4257-4267, 10.1128/iai.69.7.4257-4267.2001.
- Fulton, K.M.; Mendoza-Barberá, E.; Twine, S.M.; Tomas, J.; Merino, S.; Polar Glycosylated and Lateral Non-Glycosylated Flagella from Aeromonas hydrophila Strain AH-1 (Serotype O11). International Journal of Molecular Sciences 2015, 16, 28255-28269, 10.3390/ijms161226097.
- Canals, R.; Vilches, S.; Wilhelms, M.; Shaw, J.G.; Merino, S.; Tomás, J.M.; Non-structural flagella genes affecting both polar and lateral flagella-mediated motility in Aeromonas hydrophila. Microbiology 2007, 153, 1165-1175, 10.1099/mic.0.2006/000687-0.
- Seshadri, R.; Joseph, S.W.; Chopra, A.K.; Sha, J.; Shaw, J.; Graf, J.; Haft, D.; Wu, M.; Ren, Q.; Rosovitz, M.J.; et al. Genome Sequence of Aeromonas hydrophila ATCC 7966 T : Jack of All Trades. Journal of Bacteriology 2006, 188, 8272-8282, 10.1128/jb.00621-06.
- Forn-Cuní, G.; Tomás, J.M.; Merino, S.; Whole-Genome Sequence of Aeromonas hydrophila Strain AH-1 (Serotype O11). Genome Announcements 2016, 4, e00920-16, 10.1128/genomea.00920-16.
- Forn-Cuní, G.; Tomás, J.M.; Merino, S.; Genome Sequence of Aeromonas hydrophila Strain AH-3 (Serotype O34). Genome Announcements 2016, 4, e00919-16, 10.1128/genomea.00919-16.
- Forn-Cuní, G.; Fulton, K.M.; Smith, J.C.; Twine, S.M.; Mendoza-Barberà, E.; Tomás, J.M.; Merino, S.; Polar Flagella Glycosylation in Aeromonas: Genomic Characterization and Involvement of a Specific Glycosyltransferase (Fgi-1) in Heterogeneous Flagella Glycosylation. Frontiers in Microbiology 2021, 11, 595697, 10.3389/fmicb.2020.595697.
- Zamyatina, A.; Heine, H.; Lipopolysaccharide Recognition in the Crossroads of TLR4 and Caspase-4/11 Mediated Inflammatory Pathways. Frontiers in Immunology 2020, 11, 585146, 10.3389/fimmu.2020.585146.
- Gumenscheimer, M.; Mitov, I.; Galanos, C.; Freudenberg, M.A.; Beneficial or Deleterious Effects of a Preexisting Hypersensitivity to Bacterial Components on the Course and Outcome of Infection. Infection and Immunity 2002, 70, 5596-5603, 10.1128/iai.70.10.5596-5603.2002.
- Whitfield, C.; Valvano, M.A.; Biosynthesis and Expression of Cell-Surface Polysaccharides in Gram-Negative Bacteria. Advances in Microbial Physiology 1993, 35, 135-246, 10.1016/s0065-2911(08)60099-5.
- Silhavy, T.J.; Kahne, D.; Walker, S.; The Bacterial Cell Envelope. Cold Spring Harbor Perspectives in Biology 2010, 2, a000414, 10.1101/cshperspect.a000414.
- Lüderitz, O.; Galanos, C.; Lehmann, V.; Mayer, H.; Rietschel, E.T.; Weckesser, J.; Chemical structure and biological activities of lipid A's from various bacterial families. Naturwissenschaften 1978, 65, 578-585, 10.1007/bf00364907.
- Merino, S.; Rubires, X.; Knochel, S.; Tomas, J.M.; Emerging pathogens: Aeromonas spp.. International Journal of Food Microbiology 1995, 28, 157-168, 10.1016/0168-1605(95)00054-2.
- Wang, Z.; Li, J.; Altman, E.; Structural characterization of the lipid A region of Aeromonas salmonicida subsp. salmonicida lipopolysaccharide. Carbohydrate Research 2006, 341, 2816-2825, 10.1016/j.carres.2006.09.020.
- Hagar, J.A.; Powell, D.A.; Aachoui, Y.; Ernst, R.K.; Miao, E.A.; Cytoplasmic LPS Activates Caspase-11: Implications in TLR4-Independent Endotoxic Shock. Science 2013, 341, 1250-1253, 10.1126/science.1240988.
- Lagrange, B.; Benaoudia, S.; Wallet, P.; Magnotti, F.; Provost, A.; Michal, F.; Martin, A.; Di Lorenzo, F.; Py, B.F.; Molinaro, A.; et al. Human caspase-4 detects tetra-acylated LPS and cytosolic Francisella and functions differently from murine caspase-11. Nature Communications 2018, 9, 242, 10.1038/s41467-017-02682-y.
- Jimenez, N.; Lacasta, A.; Vilches, S.; Reyes, M.; Vazquez, J.; Aquillini, E.; Merino, S.; Regué, M.; Tomás, J.M.; Genetics and Proteomics of Aeromonas salmonicida Lipopolysaccharide Core Biosynthesis. Journal of Bacteriology 2009, 191, 2228-2236, 10.1128/jb.01395-08.
- Jimenez, N.; Canals, R.; Lacasta, A.; Kondakova, A.N.; Lindner, B.; Knirel, Y.A.; Merino, S.; Regué, M.; Tomás, J.M.; Molecular Analysis of Three Aeromonas hydrophila AH-3 (Serotype O34) Lipopolysaccharide Core Biosynthesis Gene Clusters. Journal of Bacteriology 2008, 190, 3176-3184, 10.1128/jb.01874-07.
- Wang, Z.; Li, J.; Vinogradov, E.; Altman, E.; Structural studies of the core region of Aeromonas salmonicida subsp. salmonicida lipopolysaccharide. Carbohydrate Research 2006, 341, 109-117, 10.1016/j.carres.2005.10.017.
- Knirel, Y.A.; Vinogradov, E.; Jimenez, N.; Merino, S.; Tomás, J.M.; Structural studies on the R-type lipopolysaccharide of Aeromonas hydrophila. Carbohydrate Research 2004, 339, 787-793, 10.1016/j.carres.2003.12.008.
- Raetz, C.R.H.; Whitfield, C.; Lipopolysaccharide Endotoxins. Annual Review of Biochemistry 2002, 71, 635-700, 10.1146/annurev.biochem.71.110601.135414.
- Kapp, A.; Freudenberg, M.; Galanos, C.; Induction of human granulocyte chemiluminescence by bacterial lipopolysaccharides. Infection and Immunity 1987, 55, 758-761, 10.1128/iai.55.3.758-761.1987.
- Huber, M.; Kalis, C.; Keck, S.; Jiang, Z.; Georgel, P.; Du, X.; Shamel, L.; Sovath, S.; Mudd, S.; Beutler, B.; et al. R-form LPS, the master key to the activation ofTLR4/MD-2-positive cells. European Journal of Immunology 2006, 36, 701-711, 10.1002/eji.200535593.
- Aguilar, A.; Merino, S.; Rubires, X.; Tomas, J.M.; Influence of osmolarity on lipopolysaccharides and virulence of Aeromonas hydrophila serotype O:34 strains grown at 37 degrees C. Infection and Immunity 1997, 65, 1245-1250, 10.1128/iai.65.4.1245-1250.1997.
- Merino, S.; Camprubí, S.; Tomás, J.M.; Effect of growth temperature on outer membrane components and virulence of Aeromonas hydrophila strains of serotype O:34. Infection and Immunity 1992, 60, 4343-4349, 10.1128/iai.60.10.4343-4349.1992.
- Merino, S.; Rubires, X.; Aguilar, A.; Albert, S.; Hernandez-Allé, S.; Bened, V.J.; Tomás, J.M.; Mesophilic Aeromonas sp. serogroup O:11 resistance to complement-mediated killing. Infection and Immunity 1996, 64, 5302-5309, 10.1128/iai.64.12.5302-5309.1996.
- Valvano, M. O Antigen Biosynthesis. In Comprehensive Natural Products II: Chemistry and Biology; Mander, L., Liu, H.-W., Eds.; Elsevier: Oxford, UK, 2010; pp. 297-314.
- Merino, S.; Jimenez, N.; Molero, R.; Bouamama, L.; Regué, M.; Tomás, J.M.; A UDP-HexNAc:Polyprenol-P GalNAc-1-P Transferase (WecP) Representing a New Subgroup of the Enzyme Family. Journal of Bacteriology 2011, 193, 1943-1952, 10.1128/jb.01441-10.
- Islam, S.T.; Lam, J.S.; Wzx flippase-mediated membrane translocation of sugar polymer precursors in bacteria. Environmental Microbiology 2013, 15, 1001-1015, 10.1111/j.1462-2920.2012.02890.x.
- Collins, L.V.; Hackett, J.; Molecular cloning, characterization, and nucleotide sequence of the rfc gene, which encodes an O-antigen polymerase of Salmonella typhimurium. Journal of Bacteriology 1991, 173, 2521-2529, 10.1128/jb.173.8.2521-2529.1991.
- Kalynych, S.; Valvano, M.A.; Cygler, M.; Polysaccharide co-polymerases: the enigmatic conductors of the O-antigen assembly orchestra. Protein Engineering Design and Selection 2012, 25, 797-802, 10.1093/protein/gzs075.
- Jimenez, N.; Canals, R.; Saló, M.T.; Vilches, S.; Merino, S.; Tomás, J.M.; The Aeromonas hydrophila wb * O34 Gene Cluster: Genetics and Temperature Regulation. Journal of Bacteriology 2008, 190, 4198-4209, 10.1128/jb.00153-08.
- Merino, S.; Gonzalez, V.; Tomás, J.M.; The first sugar of the repeat units is essential for the Wzy polymerase activity and elongation of the O-antigen lipopolysaccharide. Future Microbiology 2016, 11, 903-918, 10.2217/fmb-2015-0028.
- Merino, S.; Gonzalez, V.; Tomás, J.M.; The Polymerization of Aeromonas hydrophila AH-3 O-Antigen LPS: Concerted Action of WecP and Wzy. PLOS ONE 2015, 10, e0131905, 10.1371/journal.pone.0131905.
- Raetz, C.R.H. Bacterial lipopolysaccharides: A remarkable family of bioactive macroamphiphiles. In Escherichia Coli and Salmonella: Cellular and Molecular Biology; Neidhardt, F.C., Curtiss, R., III; Ingraham, J.L., Lin, E.C.C., Low, K.B., Magasanik, B., Reznikoff, W.S., Riley, M., Schaechter, M., Eds.; ASM Press: Washington, DC, USA, 1996; pp. 1035-1063.
- Thomas, L.V.; Gross, R.J.; Cheasty, T.; Rowe, B.; Extended serogrouping scheme for motile, mesophilic Aeromonas species. Journal of Clinical Microbiology 1990, 28, 980-984, 10.1128/jcm.28.5.980-984.1990.
- Merino, S.; Canals, R.; Knirel, Y.A.; Tomás, J.M.; Molecular and Chemical Analysis of the Lipopolysaccharide from Aeromonas hydrophila Strain AH-1 (Serotype O11). Marine Drugs 2015, 13, 2233-2249, 10.3390/md13042233.
- Knirel, Y.A.; Shashkov, A.S.; Senchenkova, S.N.; Merino, S.; Tomás, J.M.; Structure of the O-polysaccharide of Aeromonas hydrophila O:34; a case of random O-acetylation of 6-deoxy-L-talose. Carbohydrate Research 2002, 337, 1381-1386, 10.1016/s0008-6215(02)00136-2.
- Merino, S.; de Mendoza, E.; Canals, R.; Tomás, J.M.; Functional Genomics of the Aeromonas salmonicida Lipopolysaccharide O-Antigen and A-Layer from Typical and Atypical Strains. Marine Drugs 2015, 13, 3791-3808, 10.3390/md13063791.
- Zhang, Y.L.; Arakawa, E.; Leung, K.Y.; Novel Aeromonas hydrophila PPD134/91 Genes Involved in O-Antigen and Capsule Biosynthesis. Infection and Immunity 2002, 70, 2326-2335, 10.1128/iai.70.5.2326-2335.2002.
- Hossain, M.J.; Waldbieser, G.C.; Sun, D.; Capps, N.K.; Hemstreet, W.B.; Carlisle, K.; Griffin, M.J.; Khoo, L.; Goodwin, A.E.; Sonstegard, T.S.; et al. Implication of Lateral Genetic Transfer in the Emergence of Aeromonas hydrophila Isolates of Epidemic Outbreaks in Channel Catfish. PLOS ONE 2013, 8, e80943, 10.1371/journal.pone.0080943.
- Merino, S.; Tomás, J.M. Bacterial Capsules and Evasion of Immune Responses. In Encyclopedia of Life Sciences; John Wiley & Sons, Ltd: Chichester, UK, 2015; pp. 1-10.
- Merino, S.; Aguilar, A.; Rubires, X.; Simon-Pujol, D.; Congregado, F.; Tomás, J.M.; The role of the capsular polysaccharide of Aeromonas salmonicida in the adherence and invasion of fish cell lines. FEMS Microbiology Letters 1996, 142, 185-189, 10.1111/j.1574-6968.1996.tb08428.x.
- Merino, S.; Aguilar, A.; Rubires, X.; Abitiu, N.; Regué, M.; Tomás, J.M.; The role of the capsular polysaccharide of Aeromonas hydrophila serogroup O:34 in the adherence to and invasion of fish cell lines. Research in Microbiology 1997, 148, 625-631, 10.1016/s0923-2508(97)88086-2.
- Martínez, M.J.; Simon-Pujol, D.; Congregado, F.; Merino, S.; Rubires, X.; Tomás, J.M.; The presence of capsular polysaccharide in mesophilic Aeromonas hydrophila serotypes O:11 and O:34. FEMS Microbiology Letters 1995, 128, 69-73, 10.1111/j.1574-6968.1995.tb07502.x.
- Garrote, A.; Bonet, R.; Merino, S.; Simon-Pujol, M.D.; Congregado, F.; Occurrence of a capsule in Aeromonas salmonicida. FEMS Microbiology Letters 1992, 74, 127-131, 10.1016/0378-1097(92)90417-m.
- Wang, Z.; Larocque, S.; Vinogradov, E.; Brisson, J.R.; Dacanay, A.; Greenwell, M.; Brown, L.L.; Li, J.; Altman, E.; Structural studies of the capsular polysaccharide and lipopolysaccharide O-antigen of Aeromonas salmonicida strain 80204-1 produced under in vitro and in vivo growth conditions. European Journal of Biochemistry 2004, 271, 4507-4516, 10.1111/j.1432-1033.2004.04410.x.
- Contreras Sánchez-Matamoros, R.; Gil-Serrano, A.M.; Espuny, M.R.; Ollero, F.J.; Megías, M.; Rodríguez-Carvajal, M.A.; Structure of surface polysaccharides from Aeromonas sp. AMG272, a plant-growth promoting rhizobacterium isolated from rice rhizosphere. Carbohydrate Research 2018, 462, 1-6, 10.1016/j.carres.2018.03.012.
- Zhang, Y.L.; Lau, Y.L.; Arakawa, E.; Leung, K.Y.; Detection and genetic analysis of group II capsules in Aeromonas hydrophila. Microbiology 2003, 149, 1051-1060, 10.1099/mic.0.26144-0.
- Aguilar, A.; Merino, S.; Nogueras, M.M.; Regué, M.; Tomás, J.M.; Two genes from the capsule of Aeromonas hydrophila (serogroup O:34) confer serum resistance to Escherichia coli K12 strains. Research in Microbiology 1999, 150, 395-402, 10.1016/s0923-2508(99)80074-6.
- Preiss, J.; Romeo, T.; Molecular Biology and Regulatory Aspects of Glycogen Biosynthesis in Bacteria. Progress in Nucleic Acid Research and Molecular Biology 1994, 47, 299-329, 10.1016/s0079-6603(08)60255-x.
- Preiss, J.; Bacterial Glycogen Synthesis and its Regulation. Annual Review of Microbiology 1984, 38, 419-458, 10.1146/annurev.mi.38.100184.002223.
- Wang, L.; Wise, M.J.; Glycogen with short average chain length enhances bacterial durability. Naturwissenschaften 2011, 98, 719-729, 10.1007/s00114-011-0832-x.
- Merino, S.; Bouamama, L.; Knirel, Y.A.; Senchenkova, S.N.; Regué, M.; Tomás, J.M.; Aeromonas Surface Glucan Attached through the O-Antigen Ligase Represents a New Way to Obtain UDP-Glucose. PLoS ONE 2012, 7, e35707, 10.1371/journal.pone.0035707.
- Vilches, S.; Canals, R.; Wilhelms, M.; Saló, M.T.; Knirel, Y.A.; Vinogradog, E.; Merino, S.; Tomás, J.M.; Mesophilic Aeromonas UDP-glucose pyrophosphorylase (GalU) mutants show two types of lipopolysaccharide structures and reduced virulence. Microbiology 2007, 153, 2393-2404, 10.1099/mic.0.2007/006437-0.
- Casella, C.R.; Mitchell, T.C.; Putting endotoxin to work for us: Monophosphoryl lipid A as a safe and effective vaccine adjuvant. Cellular and Molecular Life Sciences 2008, 65, 3231-3240, 10.1007/s00018-008-8228-6.
- Zariri, A.; Pupo, E.; van Riet, E.; van Putten, J.P.M.; Modulating endotoxin activity by combinatorial bioengineering of meningococcal lipopolysaccharide. Scientific Reports 2016, 6, 36575, 10.1038/srep36575.
- Needham, B.D.; Carroll, S.M.; Giles, D.K.; Georgiou, G.; Whiteley, M.; Trent, M.S.; Modulating the innate immune response by combinatorial engineering of endotoxin. Proceedings of the National Academy of Sciences 2013, 110, 1464-1469, 10.1073/pnas.1218080110.
- Prevnar 13 . U.S. Food & Drug Administration. Retrieved 2021-12-6
- PNEUMOVAX 23—Pneumococcal Vaccine, Polyvalent . U.S. Food & Drug Administration. Retrieved 2021-12-6
- Rodríguez, I.; Chamorro, R.; Novoa, B.; Figueras, A.; β-Glucan administration enhances disease resistance and some innate immune responses in zebrafish (Danio rerio). Fish & Shellfish Immunology 2009, 27, 369-373, 10.1016/j.fsi.2009.02.007.
- Kumari, J.; Sahoo, P.K.; Dietary beta-1,3 glucan potentiates innate immunity and disease resistance of Asian catfish, Clarias batrachus (L.). Journal of Fish Diseases 2006, 29, 95-101, 10.1111/j.1365-2761.2006.00691.x.
