Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Agriculture, Dairy & Animal Science
Pentatricopeptide repeat (PPR) proteins are characterized by the presence of tandem arrays of a degenerate 35-amino-acid repeat motif, PPR motif. Based on the types of motif and their arrangement, PPR proteins are divided into two classes, P and PLS. P-class proteins only contain canonical P-motifs with 35 amino acids, whereas PLS-class proteins consist of P-, L- (35 or 36 amino acids), and S- (31 or 32 amino acids) motifs forming tandemly repeated PLS triplets.
- PPR protein
- cytoplasmic male sterility
- seed development
1. Introduction
Pentatricopeptide repeat (PPR) proteins are characterized by the presence of tandem arrays of a degenerate 35-amino-acid repeat motif, PPR motif [1]. Based on the types of motif and their arrangement, PPR proteins are divided into two classes, P and PLS. P-class proteins only contain canonical P-motifs with 35 amino acids, whereas PLS-class proteins consist of P-, L- (35 or 36 amino acids), and S- (31 or 32 amino acids) motifs forming tandemly repeated PLS triplets [2]. Many of the PLS-class proteins are carboxyl terminally extended with highly conserved E, E+, or DYW domains. Thus, PLS-class proteins can be further divided into PLS, E, E+, and DYW subclasses according to the domains identified in carboxyl terminal [3].
PPR proteins are sequence-specific RNA-binding proteins that are mostly localized to mitochondria and/or chloroplasts, where they are involved in RNA post-transcriptional processing [4] (Figure 1A). In general, the P-class PPR proteins mediate diverse aspects of RNA processing in plant organelles, while the PLS-class PPR proteins mainly function in RNA editing [5]. Mutations in these PPR protein-coding genes lead to the dysfunction of mitochondria and/or chloroplasts, thereby resulting in growth retardation, pollen abortion, and seed development defects in plants [4], indicating the important roles of PPR proteins in plant growth and development (Figure 1B).
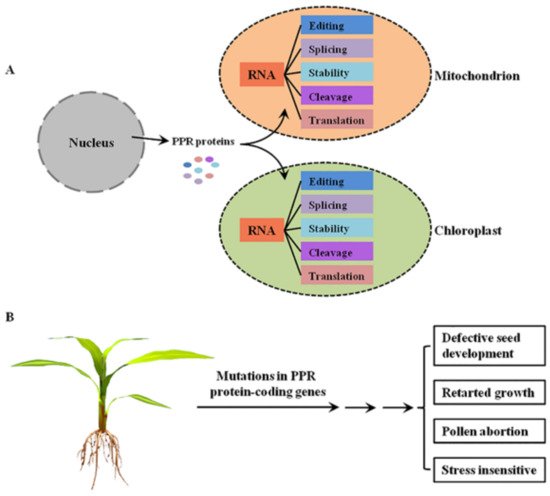
Figure 1. The functions of PPR proteins in plants. (A) The molecular functions of PPR proteins in plant mitochondria and chloroplasts. PPR proteins are encoded by nuclear genes, translated in the cytoplasm, and then imported into mitochondrion or chloroplast to mediate multiple steps of RNA processing. (B) The main growing and developmental phenotypes of plant mutants of PPR protein-coding genes.
2. Functions of PPR Proteins in Cytoplasmic Male Sterility
Cytoplasmic male sterility (CMS) is a maternally inherited trait that presents a defect in the production of viable pollen. CMS is widespread in higher plants, and has been widely used in the production of hybrid seeds and utilization of heterosis in many crop species [6,7]. Plant CMS is usually caused by mutations, rearrangements, or recombinations of mitochondrial DNA, and in many instances, male fertility can be restored specifically by restorer-of-fertility (Rf) genes in the nuclear genome [8,9]. To date, more than ten Rf genes have been cloned and functionally characterized in various crop species, and the majority of them were found to encode PPR protein, including Rf1a [10], Rf1b [11], Rf3 [12], Rf4 [13], Rf5 [14], Rf6 [15] in rice; Rfo [16], PPR-B [17], RsRf3-4 [18], Rfk1 [19] in radish; Rf1 [20], Rf2 [21] in sorghum; Rfp [22], Rfn [23] in rapeseed; Rf-PPR592 [24] in petunia; BrRfp1 [25] in Chinese cabbage; and Rfm1 [26] in barley. With the exception of the sorghum Rf1 [20] and the barely Rfm1 [26], the PPR-type Rf genes encode PPR proteins belonging to P class.
Members of P-class PPR proteins mostly function in various post-transcriptional process of organellar RNAs, such as RNA splicing, RNA stabilization, RNA cleavage, and translation [4,5]. Proteins encoded by Rf genes usually target mitochondria and act as fertility restorers by suppressing the production of mitochondrial CMS-inducing proteins [6,11]; however, the exact molecular mechanisms underlying fertility restoration by RF proteins is presently unclear. The PPR-type RF proteins have been proposed to rescue fertility by regulating the expression of CMS-conferring genes through the similar molecular mechanisms as that of other PPR proteins. In most of the CMS systems, PPR-type RF proteins bind to the mitochondrial CMS-conferring transcripts and promote their cleavage or degradation [27]. For example, rice RF1A and RF1B proteins, respectively encoded by Rf1a and Rf1b genes, have been considered to restore CMS by processing mitochondrial orf79 transcript via different mechanisms. RF1A directly binds to and cleaves the atp6-orf79 transcript at the intercistronic region, whereas RF1B promotes the rapid degradation of the atp6-orf79 transcript [10,11]. The protein encoded by rice Rf4 gene was reported to suppress WA352-mediated male sterility by reducing WA352 transcript levels [13]. Two rice RF proteins, Rfp and Rfn, are involved in transcript cleavage of orf224 and orf222, respectively [22,23].
In some CMS systems, PPR-type RF proteins restore the male fertility by impeding the translation or post-translational processing of mitochondrial CMS-inducing proteins. Studies on radish PPR-B revealed that PPR-B protein does not function through cleavage or degradation of the orf138 mRNA, but rather block its translation by inhibiting either the association with or the progression of mitochondrial ribosomes on the orf138 mRNA [17]. Similarly, rice RF3 protein does not affect the abundance of WA352 transcript but suppresses the accumulation of WA352 protein [12]. In addition, Rf1 from sorghum and Rfm1 from barely encode PPR proteins belonging to PLS class. As PLS-class PPR proteins almost exclusively play a role in RNA editing, it is possible that sorghum Rf1 and barely Rfm1 restore pollen fertility by editing S-orf transcripts or other target RNAs [8,26].
CMS was not only an ideal model system to study the interaction between mitochondrial and nuclear genomes but also a useful genetic tool for breeding to exploit hybrid vigor in crops [27]. Although PPR proteins are involved in the restoration of male fertility, functions of most PPR proteins are still obscure. Therefore, exploring the functions of PPR proteins will contribute to understanding the CMS mechanism and improving molecular breeding in crops.
3. Functions of PPR Proteins in Seed Development
In angiosperm plants, seed development starts with double fertilization of egg and central cells with two sperm cells, which leads to the formation of a diploid embryo and a triploid endosperm, and develops into mature seeds comprising three structures: maternal coat, embryo, and endosperm [49]. Development of embryo and endosperm is well correlated and regulated by numerous distinct proteins involved in many important physiological processes [50], including cell growth, RNA transcription and post-transcriptional processing, etc.
In recent years, more and more genetic and biochemical studies have shown that PPR proteins play important roles in seed development of higher plants, and loss-of-function of these PPR proteins usually leads to defects in embryogenesis and/or endosperm development [4,51]. According to the phenotypic expression, seed mutants can be divided into four major classes: empty pericarp (emp), embryo specific (emb), defective kernel (dek), and small kernel (smk). A detailed summary of the maize and Arabidopsis seed mutants caused by the functional defects of PPR proteins is provided in Table 1.
Table 1. Selected functionally characterized PPR proteins essential for seed development in maize and Arabidopsis.
| Species | Subcellular Localization |
Mutant Phenotype |
Protein Name |
PPR Class |
Function(s) | References |
|---|---|---|---|---|---|---|
| Maize | Mitochondrion | dek | DEK2 | P | RNA splicing, nad1 intron 1 | [52] |
| Mitochondrion | dek | DEK10 | PLS | RNA editing, nad3-61, 62, and cox2-550 | [53] | |
| Mitochondrion | dek | DEK35 | P | RNA splicing, nad4 intron 1 | [54] | |
| Mitochondrion | dek | DEK36 | PLS | RNA editing, atp4-59, nad7-383, and ccmFN-302 | [55] | |
| Mitochondrion | dek | DEK37 | P | RNA splicing, nad2 intron 1 | [56] | |
| Mitochondrion | dek | DEK39 | PLS | RNA editing, nad3-247 and nad3-275 | [57] | |
| Mitochondrion | dek | DEK40 | PLS | RNA editing, cox3-314, nad2-26, and nad5-1916 | [58] | |
| Mitochondrion | dek | DEK41/ DEK43 |
P | RNA splicing, nad4 intron 1 and 3 | [59,60] | |
| Mitochondrion | dek | DEK46 | PLS | RNA editing, D5-C22 of nad7 intron 3 and 4 | [61] | |
| Mitochondrion | dek | DEK53 | PLS | RNA editing, multiples sites | [62] | |
| Mitochondrion | dek | DEK55 | PLS | RNA splicing, nad4 intron 1 and 3;RNA editing, multiple sites | [63] | |
| Mitochondrion | dek | DEK605 | PLS | RNA editing, nad1-608 | [64] | |
| Mitochondrion | smk | SMK1 | PLS | RNA editing, nad7-836 | [65] | |
| Mitochondrion | smk | SMK4 | PLS | RNA editing, cox1-1489 | [66] | |
| Mitochondrion | smk | SMK6 | PLS | RNA editing, nad1-740, nad4L-110, nad7-739, and mttB-138, 139 | [67] | |
| Mitochondrion | smk | ZmSMK9 | P | RNA splicing, nad5 intron 1 and 4 | [68] | |
| Mitochondrion | smk | PPR2263 | PLS | RNA editing, nad5-1550 and cob-908 | [69] | |
| Mitochondrion | smk | MPPR6 | P | Translation, rps3 mRNA | [70] | |
| Mitochondrion | dek/smk | PPR20 | P | RNA splicing, nad2 intron 3 | [71] | |
| Mitochondrion | smk | PPR78 | P | RNA stabilization, nad5 mature mRNA | [72] | |
| Mitochondrion | emp | EMP4 | P | Expression of mitochondrial transcripts | [73] | |
| Mitochondrion | emp | EMP5 | PLS | RNA editing, multiple sites | [74] | |
| Mitochondrion | emp | EMP7 | PLS | RNA editing, ccmFN-1553 | [75] | |
| Mitochondrion | emp | EMP8 | P | RNA splicing, nad1 intron 4, nad2 intron 1, and nad4 intron 1 | [76] | |
| Mitochondrion | emp | EMP9 | PLS | RNA editing, ccmB-43 and rps4-335 | [77] | |
| Mitochondrion | emp | EMP10 | P | RNA splicing, nad2 intron 1 | [78] | |
| Mitochondrion | emp | EMP11 | P | RNA splicing, nad1 intron 1, 2, 3, and 4 | [79] | |
| Mitochondrion | emp | EMP12 | P | RNA splicing, nad2 intron 1, 2, and 4 | [80] | |
| Chloroplast | smk | qKW9 | PLS | RNA editing, NdhB-246 | [81] | |
| Mitochondrion | emp | EMP16 | P | RNA splicing, nad2 intron 4 | [82] | |
| Mitochondrion | emp | EMP17 | PLS | RNA editing, ccmFC-799 and nad2-677 | [83] | |
| Mitochondrion | emp | EMP18 | PLS | RNA editing, atp6-635 and cox2-449 | [84] | |
| Mitochondrion | emp | EMP21 | PLS | RNA editing, multiple sites | [85] | |
| Mitochondrion | emp | EMP32 | P | RNA splicing, nad7 intron 2 | [86] | |
| Mitochondrion | emp | EMP602 | P | RNA splicing, nad4 intron 1 and 3 | [87] | |
| Mitochondrion | emp | EMP603 | P | RNA splicing, nad1 intron 2 | [88] | |
| Mitochondrion | emp | PPR14 | P | RNA splicing, nad2 intron 3, nad7 intron 1 and 2 | [89] | |
| Mitochondrion | emp | PPR18 | P | RNA splicing, nad4 intron 1 | [90] | |
| Mitochondrion | emp | PPR27 | PLS | RNA editing, multiple sites | [91] | |
| Mitochondrion | emp | PPR101 | P | RNA splicing, nad5 intron 1 and 2 | [92] | |
| Mitochondrion | emp | PPR231 | P | RNA splicing, nad5 intron 1, 2, 3 and nad2 intron 3 | [92] | |
| Mitochondrion | emp | PPR-SMR1 | P | RNA splicing, multiple introns | [93] | |
| Chloroplast | emb | PPR8522 | P | RNA transcription, nearly all chloroplast-encoded genes | [94] | |
| Chloroplast | emb | EMB-7L | P | RNA splicing, multiple introns | [95] | |
| Arabidopsis | Mitochondrion | dek | OTP43 | P | RNA splicing, nad1 intron 1 | [96] |
| Mitochondrion | smk | PPR19 | P | RNA stabilization, nad1 mature mRNA | [97] | |
| Mitochondrion | emp | BLX | PLS | RNA editing, multiple sites; RNA splicing, nad1 intron 4 and nad2 intron 1 | [98] | |
| Chloroplast | emb | AtPPR2 | P | RNA translation | [99] | |
| Chloroplast | emb | ECD2 | P | RNA splicing, ndhA, ycf3 intron 1, rps12 intron 2 and clpp intron 2 | [100] | |
| Chloroplast | emb | EMB2654 | P | RNA splicing, rps12 intron 1 | [101] | |
| Mitochondrion | emb | EMB2794 | P | RNA splicing, nad2 intron 2 | [102] | |
| Nucleus | emb | GRP23 | P | RNA transcription | [103] | |
| Mitochondrion | emb | MID1 | P | RNA splicing, nad2 intron 1 | [104] | |
| Chloroplast | emb | PMD3 | P | RNA splicing, trnA, ndhB, and clpP-1 | [105] |
Most PPR proteins identified to date are targeted to mitochondria and/or chloroplasts [4]. The disruption of PPR proteins localized to chloroplasts usually results in the emb mutants that are defective in embryogenesis, but relatively normal in endosperm development. For instance, PPR8522 [94] and EMB-7L [95] in maize and GRP23 [103] in Arabidopsis are necessary for embryogenesis, and their mutations lead to arrested embryo development at the transition stage, resulting in an embryo-lethal phenotype. For mitochondrion-targeted PPR proteins, their disruptions mostly cause diverse seed development mutants, including smk, dek, and emp, with different degrees of defects in embryo and endosperm. The loss-of-function of SMK1 [65], SMK4 [66], SMK6 [67], ZmSMK9 [68], PPR2263 [69], and MPPR6 [70] in maize and PPR19 [97] in Arabidopsis arrests both embryo and endosperm development, resulting in smk phenotypes. Some characterized mitochondrion-targeted PPR proteins, such as DEK2 [52], DEK10 [53], DEK35 [54], DEK36 [55], DEK37 [56], DEK39 [57], DEK40 [58], DEK41/43 [59,60], DEK46 [61], and DEK53 [62] in maize, are necessary for seed development, and their disruptions result in dek mutants with arrested development of both the embryo and endosperm at an early stage. Meanwhile, many PPR proteins are targeted to mitochondria and function in development of both embryo and endosperm, and mutations in their encoding genes arrest embryo and endosperm development at early stages and even result in embryo lethality. For example, Emp5 encodes a mitochondrion-targeted DYW-type PPR protein, the emp5 mutants exhibit abortion of embryo and endosperm development at early stages in maize [74]. Loss-of-function of the mitochondrial P-type PPR protein EMP10 severely disturbs embryo and endosperm development, resulting in empty pericarp or papery seeds in maize [78]. Additionally, the P-type protein PPR5 was recently identified as a regulator required for endosperm development in rice, the ppr5 mutants develop small starch grains [106].
This entry is adapted from the peer-reviewed paper 10.3390/ijms222011274
This entry is offline, you can click here to edit this entry!
