Antimicrobial resistance (AMR) is one of the top 10 global public health threats facing humanity, especially in low-resource settings, and requires an interdisciplinary response across academia, government, countries, and societies. If unchecked, AMR will hamper progress towards reaching the United Nations Sustainable Development Goals (SDGs), including ending poverty and hunger, promoting healthy lives and well-being, and achieving sustained economic growth. There are many global initiatives to curb the effects of AMR, but significant gaps remain. New ways of thinking and operating in the context of the SDGs are essential to making progress. In this entry, we define the next generation of the AMR research network, its composition, and strategic activities that can help mitigate the threats due to AMR at the local, regional, and global levels. This is supported by a review of recent literature and bibliometric and network analyses to examine the current and future state of AMR research networks for global health and sustainable development.
- AMR
- networks
- bibliometrics
- SDGs
1. Introduction
2. Drivers of Antimicrobial Resistance
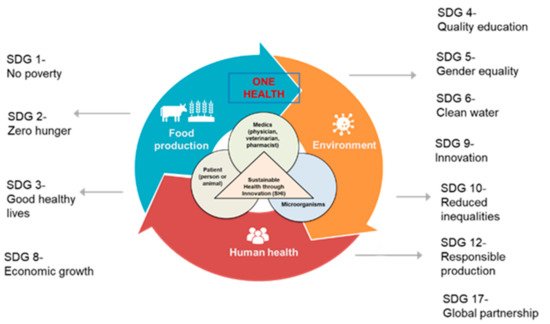
3. AMR and Sustainable Development Goals
4. Global and National AMR Initiatives
4.1. Global Health
4.2. Global Animal Health
4.3. Diagnostics: From Biomarkers to Diseases
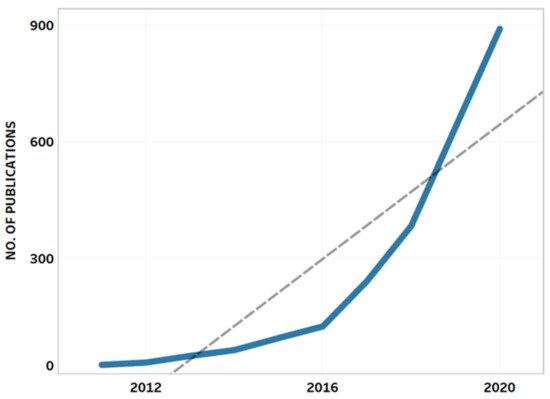
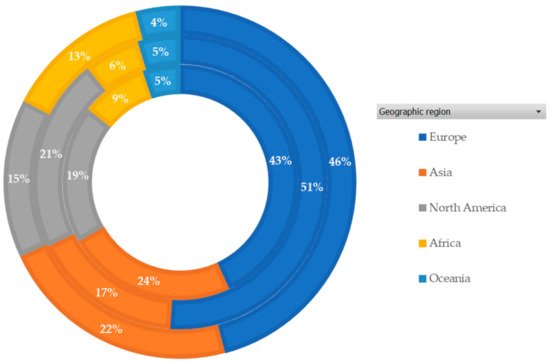
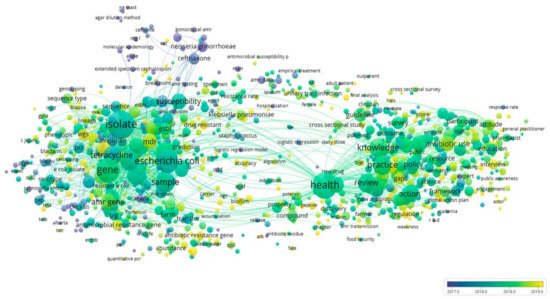
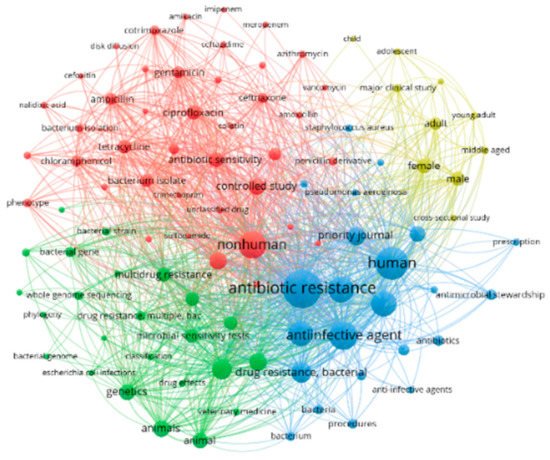
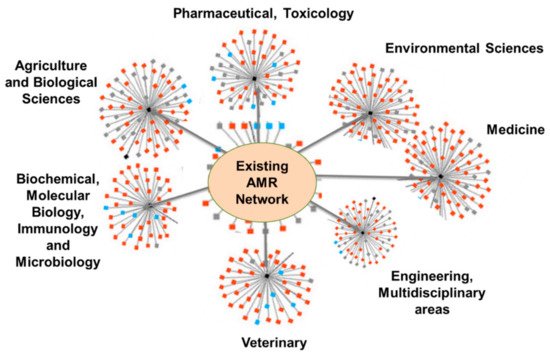
4.4. Bibliometric Analysis and AMR Global Research Outputs
Review of Bibliometrics and AMR Research
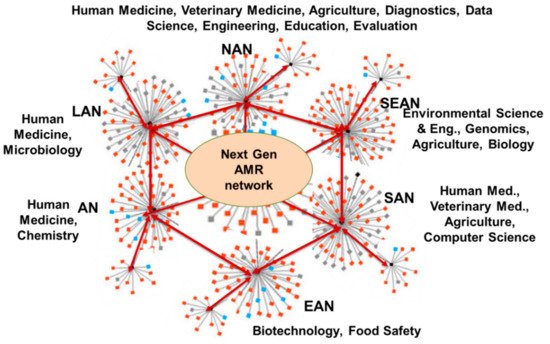
5. Defining the Next Generation of AMR Network
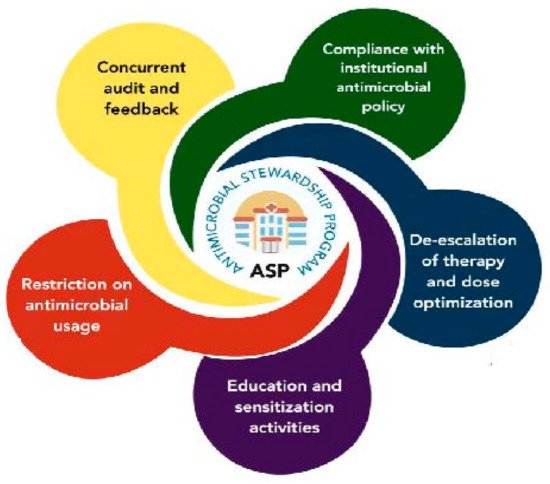
5.1. Antimicrobial Stewardship
5.2. Data-Based Tools and Mobile Applications
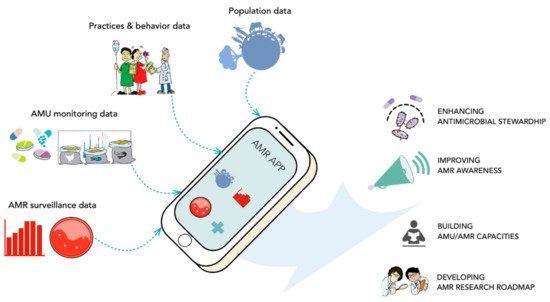
5.3. Training and Network Linkages
6. Conclusions and Prospects
References
- WHO. Antimicrobial Resistance. Available online: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (accessed on 25 March 2021).
- Jonas, O.B.; Irwin, A.; Berthe, F.C.J.; Le Gall, F.G.; Marquez, P.V. Drug-resistant infections: A Threat to Our Economic Future (Vol. 2): Final Report. Available online: https://documents.worldbank.org/en/publication/documents-reports/documentdetail/323311493396993758/final-report (accessed on 17 May 2021).
- Kirchhelle, C.; Atkinson, P.; Broom, A.; Chuengsatiansup, K.; Ferreira, J.P.; Fortané, N.; Frost, I.; Gradmann, C.; Hinchliffe, S.; Hoffman, S.J.; et al. Setting the Standard: Multidisciplinary Hallmarks for Structural, Equitable and Tracked Antibiotic Policy. BMJ Glob. Health 2020, 5, e003091.
- UN. Take Action for the Sustainable Development Goals—United Nations Sustainable Development. Available online: https://www.un.org/sustainabledevelopment/sustainable-development-goals/ (accessed on 16 May 2021).
- NAP. Download: Combating Antimicrobial Resistance: A One Health Approach to a Global Threat: Proceedings of a Workshop|The National Academies Press. Available online: https://www.nap.edu/download/24914 (accessed on 25 March 2021).
- Infection Control Today. Antimicrobial Resistance: Stewardship and Strategies for Conquering a Global Threat. Available online: https://www.infectioncontroltoday.com/view/antimicrobial-resistance-stewardship-and-strategies-conquering-global (accessed on 5 April 2021).
- CDC. Antibiotic-Resistant Germs: New Threats. Available online: https://www.cdc.gov/drugresistance/biggest-threats.html (accessed on 25 March 2021).
- CDC Online Newsroom. Press Release; Untreatable: Report by CDC Details Todays Drug-Resistant Health Threats. Available online: https://www.cdc.gov/media/releases/2013/p0916-untreatable.html (accessed on 25 March 2021).
- Wellcome Trust. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations/the Review on Antimicrobial Resistance Chaired by Jim O’Neill. Available online: https://wellcomecollection.org/works/thvwsuba (accessed on 25 March 2021).
- WHO. Antimicrobial Resistance in the Food Chain. Available online: http://www.who.int/foodsafety/areas_work/antimicrobial-resistance/amrfoodchain/en/ (accessed on 5 May 2021).
- Grøntvedt, C.A.; Elstrøm, P.; Stegger, M.; Skov, R.L.; Skytt Andersen, P.; Larssen, K.W.; Urdahl, A.M.; Angen, Ø.; Larsen, J.; Åmdal, S.; et al. Methicillin-Resistant Staphylococcus Aureus CC398 in Humans and Pigs in Norway: A “One Health” Perspective on Introduction and Transmission. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2016, 63, 1431–1438.
- Price, L.B.; Stegger, M.; Hasman, H.; Aziz, M.; Larsen, J.; Andersen, P.; Pearson, T.; Waters, A.; Foster, J.; Schupp, J.; et al. mBio. Staphylococcus aureus CC398: Host Adaptation and Emergence of Methicillin Resistance in Livestock. Available online: https://mbio.asm.org/content/3/1/e00305-11 (accessed on 5 April 2021).
- Ewers, C.; Bethe, A.; Semmler, T.; Guenther, S.; Wieler, L.H. Extended-Spectrum β-Lactamase-Producing and AmpC-Producing Escherichia coli from Livestock and Companion Animals, and Their Putative Impact on Public Health: A Global Perspective. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2012, 18, 646–655.
- Al Bayssari, C.; Dabboussi, F.; Hamze, M.; Rolain, J.-M. Emergence of Carbapenemase-Producing Pseudomonas Aeruginosa and Acinetobacter Baumannii in Livestock Animals in Lebanon. J. Antimicrob. Chemother. 2015, 70, 950–951.
- Fletcher, S. Understanding the Contribution of Environmental Factors in the Spread of Antimicrobial Resistance. Environ. Health Prev. Med. 2015, 20, 243–252.
- Lerminiaux, N.A.; Cameron, A.D.S. Horizontal Transfer of Antibiotic Resistance Genes in Clinical Environments. Can. J. Microbiol. 2019, 65, 34–44.
- Oladeinde, A.; Cook, K.; Lakin, S.M.; Woyda, R.; Abdo, Z.; Looft, T.; Herrington, K.; Zock, G.; Lawrence, J.P.; Thomas, J.C.; et al. Horizontal Gene Transfer and Acquired Antibiotic Resistance in Salmonella Enterica Serovar Heidelberg Following In Vitro Incubation in Broiler Ceca. Appl. Environ. Microbiol. 2019, 85, e01903-19.
- World Bank. Pulling Together to Beat Superbugs: Knowledge and Implementation Gaps in Addressing Antimicrobial Resistance. Available online: https://www.worldbank.org/en/topic/agriculture/publication/pulling-together-to-beat-superbugs-knowledge-and-implementation-gaps-in-addressing-antimicrobial-resistance (accessed on 25 March 2021).
- Kearns, J. The role of chemical exposures in reducing the effectiveness of water–sanitation–hygiene interventions in Bangladesh, Kenya, and Zimbabwe. WIREs Water 2020, 7, e1478.
- Giowanella, M.; Bozza, A.; do Rocio Dalzoto, P.; Dionísio, J.A.; Andraus, S.; Guimarães, E.L.G.; Pimentel, I.C. Microbiological Quality of Water from the Rivers of Curitiba, Paraná State, Brazil, and the Susceptibility to Antimicrobial Drugs and Pathogenicity of Escherichia coli. Environ. Monit. Assess. 2015, 187, 673.
- IWA Publishing. Bacteriological and Physico-Chemical Quality Assessment of Household Drinking Water in Ado-Ekiti, Nigeria|Water Supply. Available online: https://iwaponline.com/ws/article/11/1/79/25143/Bacteriological-and-physico-chemical-quality (accessed on 5 May 2021).
- Blom, K. Drainage Systems, an Occluded Source of Sanitation Related Outbreaks. Arch. Public Health 2015, 73, 8.
- Baghel, V.S.; Singh, J.; Gopal, K. Antimicrobial Resistance among Enteric Bacteria, Isolated from Runoff of the Gangotri Glacier, Western Himalaya India. J. Environ. Biol. 2003, 24, 349–356.
- Singh, A.K.; Das, S.; Singh, S.; Gajamer, V.R.; Pradhan, N.; Lepcha, Y.D.; Tiwari, H.K. Prevalence of Antibiotic Resistance in Commensal Escherichia coli among the Children in Rural Hill Communities of Northeast India. PLoS ONE 2018, 13, e0199179.
- Alders, R.G.; Dumas, S.E.; Rukambile, E.; Magoke, G.; Maulaga, W.; Jong, J.; Costa, R. Family Poultry: Multiple Roles, Systems, Challenges, and Options for Sustainable Contributions to Household Nutrition Security through a Planetary Health Lens. Matern. Child. Nutr. 2018, 14 (Suppl. S3), e12668.
- Acharya, K.P.; Wilson, R.T. Antimicrobial Resistance in Nepal. Front. Med. 2019, 6, 1–10.
- Van Boeckel, T.; Gandra, S.; Ashok, A.; Caudron, Q.; Grenfell, B.; Levin, S.; Laxminarayan, R. Global Antibiotic Consumption 2000 to 2010: An Analysis of National Pharmaceutical Sales Data. Lancet Infect. Dis. 2014, 14, 742–750.
- SDG Knowledge Hub|Guest Article: Tracking Antimicrobial Resistance in the Sustainable Development Goal. Available online: https://sdg.iisd.org/commentary/guest-articles/tracking-antimicrobial-resistance-in-the-sustainable-development-goals/ (accessed on 7 May 2021).
- UN|Department of Economic and Social Affairs|Sustainable Development.
- Cars, O.; Jasovsky, D. Antibiotic Resistance (ABR)—No Sustainability without Antibiotics. Available online: https://sustainabledevelopment.un.org/content/documents/634484-Cars-Antibiotic%20resistance%20(ABR)%20-%20No%20sustainability%20without%20antibiotics%20-%20ReAct.pdf (accessed on 15 May 2021).
- Cars, O.; Murray, M.; Nordberg, O.; Lundborg, C. Meeting the Challenge of Antibiotic Resistance. BMJ 2008, 337, a1438.
- Nathan, C.; Cars, O. Antibiotic Resistance—Problems, Progress, and Prospects. N. Engl. J Med. 2014, 371, 1761–1763.
- McAdams, D.; Waldetoft, K.; Tedijanto, C.; Brown, S. Resistance Diagnostics as a Public Health Tool to Combat Antibiotic Resistance: A Model-Based Evaluation. PLoS Biol. 2019, 17, e3000250.
- Gordillo-Marroquín, C.; Gómez-Velasco, A.; Sánchez-Pérez, H.J.; Pryg, K.; Shinners, J.; Murray, N.; Muñoz-Jiménez, S.G.; Bencomo-Alerm, A.; Gómez-Bustamante, A.; Jonapá-Gómez, L.; et al. Magnetic Nanoparticle-Based Biosensing Assay Quantitatively Enhances Acid-Fast Bacilli Count in Paucibacillary Pulmonary Tuberculosis. Biosensors 2018, 8, 128.
- Zeeshan, K.; Daya, P.; Tirumalai, S.; Alocilja, E. Impedance and Magnetohydrodynamic Measurements for Label Free Detection and Differentiation of E. coli and S. Aureus Using Magnetic Nanoparticles. IEEE Trans. NanoBiosci. 2018, 17, 443–448.
- Etyang, A.O.; Scott, J.A.G. Medical Causes of Admissions to Hospital among Adults in Africa: A Systematic Review. Glob. Health Action 2013, 6, 1–14.
- WHO. Global Strategy for Containment of Antimicrobial Resistance. Available online: https://www.who.int/drugresistance/WHO_Global_Strategy_English.pdf (accessed on 2 April 2021).
- Jasovský, D.; Littmann, J.; Zorzet, A.; Cars, O. Antimicrobial Resistance—A Threat to the World’s Sustainable Development. Ups. J. Med. Sci. 2016, 121, 159–164.
- WHO. Global Action Plan on Antimicrobial Resistance. Available online: http://www.who.int/antimicrobial-resistance/publications/global-action-plan/en/ (accessed on 2 April 2021).
- FAO. The FAO Action Plan on Antimicrobial Resistance 2016–2020. Available online: http://www.fao.org/3/i5996e/i5996e.pdf (accessed on 2 April 2021).
- World Organisation for Animal Health; OIE. The OIE Strategy on Antimicrobial Resistance and Prudent Use of Antimicrobials. Available online: https://www.oie.int/app/uploads/2021/03/en-oie-amrstrategy.pdf. (accessed on 7 June 2021).
- CDC. Tracking Threats Using Data. Available online: https://www.cdc.gov/drugresistance/biggest-threats/tracking.html (accessed on 26 March 2021).
- GLASS. Global Antimicrobial Resistance Surveillance System (GLASS). Available online: http://www.who.int/glass/en/ (accessed on 26 March 2021).
- GLASS. Global Antimicrobial Resistance and Use Surveillance System (GLASS) Report. Available online: http://www.who.int/glass/resources/publications/early-implementation-report-2020/en/ (accessed on 18 May 2021).
- European Centre for Disease Prevention and Control. European Surveillance of Antimicrobial Consumption Network (ESAC-Net). Available online: https://www.ecdc.europa.eu/en/about-us/partnerships-and-networks/disease-and-laboratory-networks/esac-net (accessed on 26 March 2021).
- WHO. One Health. Available online: https://www.who.int/news-room/q-a-detail/one-health (accessed on 19 December 2020).
- WHO. OneHealth: OIE—World Organisation for Animal Health. Available online: https://www.oie.int/en/for-the-media/onehealth/ (accessed on 19 December 2020).
- JPIAMR Secretariat. JPIAMR Launches AMR Research Funding Dashboard. Available online: https://www.jpiamr.eu/new-mapping-of-amr-research-funding-published-by-jpiamr/ (accessed on 19 December 2020).
- Candid. Antimicrobial Resistance Fund Launched with $1 Billion in Investments. Available online: http://philanthropynewsdigest.org/news/antimicrobial-resistance-fund-launched-with-1-billion-in-investments (accessed on 5 April 2021).
- Ruegg, University of Wisconsin-Madison, USA; Taylor & Francis Group. Minimizing the Development of Antimicrobial Resistance on Dairy Farms: Appropriate Use of Antibiotics for the Treatment of Mastitis Pamela L. Available online: https://www.taylorfrancis.com/chapters/edit/10.4324/9781351114202-10/minimizing-development-antimicrobial-resistance-dairy-farms-appropriate-use-antibiotics-treatment-mastitis-pamela-ruegg-university-wisconsin-madison-usa (accessed on 29 April 2021).
- Laxminarayan, R.; Matsoso, P.; Pant, S.; Brower, C.; Røttingen, J.-A.; Klugman, K.; Davies, S. Access to Effective Antimicrobials: A Worldwide Challenge. Lancet 2016, 387, 168–175.
- WHO. OneHealth: OIE—World Organisation for Animal Health. Available online: https://www.oie.int/en/what-we-do/global-initiatives/antimicrobial-resistance/#ui-id-3 (accessed on 19 August 2021).
- Schrag, N.F.D.; Apley, M.D.; Godden, S.M.; Lubbers, B.V.; Singer, R.S. Antimicrobial Use Quantification in Adult Dairy Cows—Part 1—Standardized Regimens as a Method for Describing Antimicrobial Use. Zoonoses Public Health 2020, 67 (Suppl. S1), 51–68.
- Schrag, N.; Apley, M.D.; Godden, S.M.; Singer, R.; Lubbers, B. Antimicrobial Use Quantification in Adult Dairy Cows—Part 2—Developing a Foundation for Pharmacoepidemiology by Comparing Measurement Methods. Zoonoses Public Health 2020, 67 (Suppl. S1), 67–81.
- Schrag, N.; Apley, M.D.; Godden, S.M.; Apley, M.D.; Singer, R.; Lubbers, B. Antimicrobial Use Quantification in Adult Dairy Cows—Part 3—Use measured by Standardized Regimens and Grams on 29 Dairies in the United States. Zoonoses Public Health 2020, 67 (Suppl. S1), 82–93.
- Hope, K.J.; Apley, M.D.; Schrag, N.F.D.; Lubbers, B.V.; Singer, R.S. Antimicrobial Use in 22 U.S. Beef Feedyards: 2016–2017. Zoonoses Public Health 2020, 67 (Suppl. S1), 94–110.
- Hope, K.J.; Apley, M.D.; Schrag, N.F.D.; Lubbers, B.V.; Singer, R.S. Comparison of Surveys and Use Records for Quantifying Medically Important Antimicrobial Use in 18 U.S. Beef Feedyards. Zoonoses Public Health 2020, 67 (Suppl. S1), 111–123.
- Singer, R.S.; Porter, L.J.; Schrag, N.F.D.; Davies, P.R.; Apley, M.D.; Bjork, K. Estimates of On-Farm Antimicrobial Usage in Turkey Production in the United States, 2013–2017. Zoonoses Public Health 2020, 67 (Suppl. S1), 36–50.
- De Campos, J.L.; Kates, A.; Steinberger, A.; Sethi, A.; Suen, G.; Shutske, J.; Safdar, N.; Goldberg, T.; Ruegg, P.L. Quantification of Antimicrobial Usage in Adult Cows and Preweaned Calves on 40 Large Wisconsin Dairy Farms Using Dose-Based and Mass-Based Metrics. J. Dairy Sci. 2021, 104, 4727–4745.
- Singer, R.; Porter, L.J.; Schrag, N.; Davies, P.R.; Apley, M.D.; Bjork, K.E. Estimates of On-farm Antimicrobial Usage in Broiler Chicken Production in the United States, 2013–2017. Zoonoses Public Health 2020, 67 (Suppl. S1), 22–35.
- Kuipers, A.; Koops, W.J.; Wemmenhove, H. Antibiotic Use in Dairy Herds in the Netherlands from 2005 to 2012. J. Dairy Sci. 2016, 99, 1632–1648.
- Morgan, V.; Casso-Hartmann, L.; Bahamon-Pinzon, D.; McCourt, K.; Hjort, R.G.; Bahramzadeh, S.; Velez-Torres, I.; McLamore, E.; Gomes, C.; Alocilja, E.C.; et al. Sensor-as-a-Service: Convergence of Sensor Analytic Point Solutions (SNAPS) and Pay-A-Penny-Per-Use (PAPPU) Paradigm as a Catalyst for Democratization of Healthcare in Underserved Communities. Diagnostics 2020, 10, 22.
- McLamore, E.S.; Palit Austin Datta, S.; Morgan, V.; Cavallaro, N.; Kiker, G.; Jenkins, D.M.; Rong, Y.; Gomes, C.; Claussen, J.; Vanegas, D.; et al. SNAPS: Sensor Analytics Point Solutions for Detection and Decision Support Systems. Sensors 2019, 19, 4935.
- Cleland, D.A.; Eranki, A.P. Procalcitonin; StatPearls Publishing: Treasure Island, FL, USA, 2021.
- Sawatzky, P.; Liu, G.; Dillon, J.-A.R.; Allen, V.; Lefebvre, B.; Hoang, L.; Tyrrell, G.; Van Caeseele, P.; Levett, P.; Martin, I. Quality Assurance for Antimicrobial Susceptibility Testing of Neisseria Gonorrhoeae in Canada, 2003 to 2012. J. Clin. Microbiol. 2015, 53, 3646–3649.
- van Belkum, A.; Bachmann, T.T.; Lüdke, G.; Lisby, J.G.; Kahlmeter, G.; Mohess, A.; Becker, K.; Hays, J.P.; Woodford, N.; Mitsakakis, K.; et al. Developmental Roadmap for Antimicrobial Susceptibility Testing Systems. Nat. Rev. Microbiol. 2019, 17, 51–62.
- Vasala, A.; Hytönen, V.P.; Laitinen, O.H. Modern Tools for Rapid Diagnostics of Antimicrobial Resistance. Front. Cell. Infect. Microbiol. 2020, 10.
- Davies, J.; Davies, D. Origins and Evolution of Antibiotic Resistance. Microbiol. Mol. Biol. Rev. 2010, 74, 417–433.
- OECD. OECD Glossary of Statistical Terms—Bibliometrics Definition. Available online: https://stats.oecd.org/glossary/detail.asp?ID=198 (accessed on 30 March 2021).
- Sweileh, W.M.; Shraim, N.Y.; Al-Jabi, S.W.; Sawalha, A.F.; AbuTaha, A.S.; Zyoud, S.H. Bibliometric Analysis of Global Scientific Research on Carbapenem Resistance (1986–2015). Ann. Clin. Microbiol. Antimicrob. 2016, 15, 1–11.
- Torres, R.T.; Carvalho, J.; Cunha, M.V.; Serrano, E.; Palmeira, J.D.; Fonseca, C. Temporal and Geographical Research Trends of Antimicrobial Resistance in Wildlife—A Bibliometric Analysis. One Health 2020, 11, 100198.
- Sweileh, W.M.; Moh’d Mansour, A. Bibliometric Analysis of Global Research Output on Antimicrobial Resistance in the Environment (2000–2019). Glob. Health Res. Policy 2020, 5, 37.
- Verma, M.; Shafiq, N.; Tripathy, J.P.; Nagaraja, S.B.; Kathirvel, S.; Chouhan, D.K.; Arora, P.; Singh, T.; Jain, K.; Gautam, V.; et al. Antimicrobial Stewardship Programme in a Trauma Centre of a Tertiary Care Hospital in North India: Effects and Implementation Challenges. J. Glob. Antimicrob. Resist. 2019, 17, 283–290.
- Antimicrobial Stewardship Programmes in Health-Care Facilities in Low- and Middle-Income Countries: A WHO Practical Toolkit. JAC Antimicrob. Resist. 2019, 1, dlz072.
- Liaskou, M.; Duggan, C.; Joynes, R.; Rosado, H.R. Pharmacy’s Role in Antimicrobial Resistance and Stewardship. Available online: https://pharmaceutical-journal.com/article/research/pharmacys-role-in-antimicrobial-resistance-and-stewardship (accessed on 20 April 2021).
- Davey, P.; Pagliari, C.; Hayes, A. The Patient’s Role in the Spread and Control of Bacterial Resistance to Antibiotics. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2002, 8 (Suppl. S2), 43–68.
- Van den Bogaard, A.E.; Stobberingh, E.E. Antibiotic Usage in Animals: Impact on Bacterial Resistance and Public Health. Drugs 1999, 58, 589–607.
- Patel, S.J.; Wellington, M.; Shah, R.M.; Ferreira, M.J. Antibiotic Stewardship in Food-Producing Animals: Challenges, Progress, and Opportunities. Clin. Ther. 2020, 42, 1649–1658.
- Tao, W.; Zhang, X.-X.; Zhao, F.; Huang, K.; Ma, H.; Wang, Z.; Ye, L.; Ren, H. High Levels of Antibiotic Resistance Genes and Their Correlations with Bacterial Community and Mobile Genetic Elements in Pharmaceutical Wastewater Treatment Bioreactors. PLoS ONE 2016, 11, e0156854.
- Zhou, Z.; Zheng, J.; Wei, Y.; Chen, T.; Dahlgren, R.; Shang, X.; Chen, H. Antibiotic Resistance Genes in an Urban River as Impacted by Bacterial Community and Physicochemical Parameters. Environ. Sci. Pollut Res. Int. 2017, 24, 23753–23762.
- Xiong, W.; Sun, Y.; Zhang, T.; Ding, X.; Li, Y.; Wang, M.; Zeng, Z. Antibiotics, Antibiotic Resistance Genes, and Bacterial Community Composition in Fresh Water Aquaculture Environment in China. Microb. Ecol. 2015, 70, 425–432.
- Wang, M.; Liu, P.; Xiong, W.; Zhou, Q.; Wangxiao, J.; Zeng, Z.; Sun, Y. Fate of Potential Indicator Antimicrobial Resistance Genes (ARGs) and Bacterial Community Diversity in Simulated Manure-Soil Microcosms. Ecotoxicol. Environ. Saf. 2018, 147, 817–823.
- Marti, E.; Jofre, J.; Balcazar, J.L. Prevalence of Antibiotic Resistance Genes and Bacterial Community Composition in a River Influenced by a Wastewater Treatment Plant. PLoS ONE 2013, 8, e78906.
- Szekeres, E.; Baricz, A.; Chiriac, C.M.; Farkas, A.; Opris, O.; Soran, M.-L.; Andrei, A.-S.; Rudi, K.; Balcázar, J.L.; Dragos, N.; et al. Abundance of Antibiotics, Antibiotic Resistance Genes and Bacterial Community Composition in Wastewater Effluents from Different Romanian Hospitals. Environ. Pollut. 2017, 225, 304–315.
- Punina, N.V.; Makridakis, N.M.; Remnev, M.A.; Topunov, A.F. Whole-Genome Sequencing Targets Drug-Resistant Bacterial Infections. Hum. Genomics 2015, 9, 19.
- Schürch, A.C.; van Schaik, W. Challenges and Opportunities for Whole-Genome Sequencing-Based Surveillance of Antibiotic Resistance. Ann. N. Y. Acad. Sci. 2017, 1388, 108–120.
- Oniciuc, E.A.; Likotrafiti, E.; Alvarez-Molina, A.; Prieto, M.; Santos, J.A.; Alvarez-Ordóñez, A. The Present and Future of Whole Genome Sequencing (WGS) and Whole Metagenome Sequencing (WMS) for Surveillance of Antimicrobial Resistant Microorganisms and Antimicrobial Resistance Genes across the Food Chain. Genes 2018, 9, 268.
- Tyler, A.D.; Mataseje, L.; Urfano, C.J.; Schmidt, L.; Antonation, K.S.; Mulvey, M.R.; Corbett, C.R. Evaluation of Oxford Nanopore’s MinION Sequencing Device for Microbial Whole Genome Sequencing Applications. Sci. Rep. 2018, 8, 10931.
- Brown, B.L.; Watson, M.; Minot, S.S.; Rivera, M.C.; Franklin, R.B. MinIONTM Nanopore Sequencing of Environmental Metagenomes: A Synthetic Approach. GigaScience 2017, 6, 1–10.
- Babalola, O.O.; Fadiji, A.E.; Ayangbenro, A.S. Shotgun Metagenomic Data of Root Endophytic Microbiome of Maize (Zea mays L.). Data Brief 2020, 31, 105893.
- Chowdhury, A.; Lofgren, E.; Moehring, R.; Broschat, S. Identifying Predictors of Antimicrobial Exposure in Hospitalized Patients using a Machine Learning Approach. J. Appl. Microbiol. 2020, 128, 688–696.
- Weinstein, Z.B.; Bender, A.; Cokol, M. Prediction of Synergistic Drug Combinations. Curr. Opin. Syst. Biol. 2017, 4, 24–28.
- Rodríguez-González, A.; Zanin, M.; Menasalvas-Ruiz, E. Public Health and Epidemiology Informatics: Can Artificial Intelligence Help Future Global Challenges? An Overview of Antimicrobial Resistance and Impact of Climate Change in Disease Epidemiology. Yearb. Med. Inform. 2019, 28, 224–231.
- Briliyanti, A.; Rojewski, J.; Luchini-Colbry, K.; Colbry, D. CyberAmbassadors: Results from Pilot Testing a New Professional Skills Curriculum. Pract. Exp. Adv. Res. Comput. 2020, 379–385.
