Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is an old version of this entry, which may differ significantly from the current revision.
Subjects:
Engineering, Biomedical
The MYB transcription factors (TFs) are evolving as critical role in the regulation of the phenylpropanoid and tanshinones biosynthetic pathway. MYB TFs relate to a very important gene family, which are involved in the regulation of primary and secondary metabolisms, terpenoids, bioactive compounds, plant defense against various stresses and cell morphology.
- tanshinones
- phenolic acid
- plant defense
- pdLNLD/ELxiG/S motif
- flavonoids
- repressor MYB
- bioactive compounds
1. Introduction
The compounds derived from phenylpropanoid denote a different class of secondary metabolites, which start from key enzyme phenylalanine. Phenylpropanoid derived metabolites play an important function in plant resistance mechanisms against biotic and abiotic stress, regulate plant growth and development [1,2] and male fertility [3]. Several of these phenylpropanoid derived compounds are considered to be valuable to human welfare and health. MYB protein associated with a big class of transcription factors, which are responsible for the regulation of the biosynthetic pathway of phenylpropanoid resulting compounds [4]. In plants, phenylpropanoid derived secondary metabolites mainly consist of flavonoid, monolignol, stilbenes, terpenoids and different phenolic acid. Many of these compounds play a key role in identified plants, including as UV light protectants, phytoalexins, carotenoids, strengthen the cell wall and signaling molecules [5].
The pigments that provide the different colors to vegetables, fruits, ornamental foliage, leaves, ornamental flowers and seeds are called flavonoids, which provide health benefits to the human and animals [6]. Flavonoids are the secondary metabolites that are broadly distributed in the plant kingdom, which play key roles in plant defense and development. These secondary metabolites can be divided into different groups based on differences in their structure, such as anthocyanin, proanthocyanin, chalcones, flavones, flavonols, flavandiols, isoflavonoids and phlobaphenes [7]. Flavonoids are the most common occurring pigment in plants. Anthocyanins are commonly known as flavonoid compounds providing blues, pink hues, orange, yellow and red colors to flowers, fruits and vegetables. Anthocyanins play very significant physiological and ecological roles in plants. Anthocyanin is most noticeable in young leaves where they defend developing tissues from light stress. Anthocyanins play a key role in seed dispersal and pollination by attracting the pollinator agent in mature fruits and flowers. Proanthocyanin (also known as tannins) provides as significantly roles, such as strengthening the seed coat and stress tolerance in plants [8]. Furthermore, these compounds are concerned in the regulation signaling, when legume are in nodulation process, transportation of auxin and male fertility determination. Moreover, these compounds are involved in plant defense in opposition to stress (biotic and abiotic). These compounds have very imperative values as nutritional and pharmaceutical compounds [9].
Phenylalanine ammonia-lyase (PAL), cinnamate 4-hydroxylase (C4H), and p-coumaroyl coenzyme A ligase (4CL) are very important key enzymes, which jointly catalyzed stages (First three stages) involved in biosynthesis of compounds, which are derived from phenylpropanoid, as shown in (Figure 1).
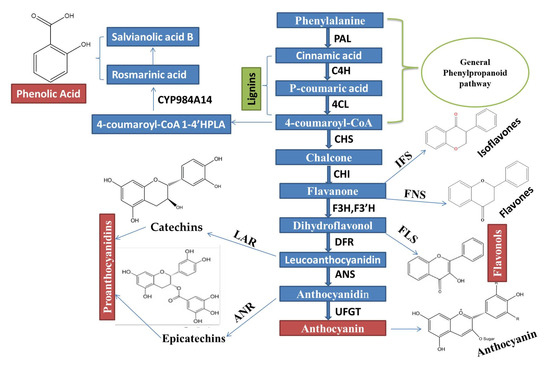
Figure 1. An Integrated regulatory mechanisms network of the Phenylpropanoid biosynthetic pathway, which is controlled through MYB TFs and MBW complex activation, is constructed based on the recent remarkable research advancement.
Various transcription factors (TFs), including R2R3 MYB, WD40 repeat (WDR) proteins and basic helix-loop-helix (bHLH) and control the biosynthesis of flavonoid compounds [10]. A complex of MYB-bHLH-WDR (MBW) shows action, in order to trigger the structural genes responsible for the process of flavonoid biosynthesis. In several plants, including Helianthus annuus L., Arabodopsis thialana, Mimulus guttatus, Camellia sinensis, Narcissus tazetta. L. Narcissus tazetta, Zea mays, Glycine max, Medicago truncatula, Fragaria × ananassa, populous, Petunia x hybrida, Malus domestica, and Vitis vinifera L., these transcription factors have been functionally characterized well [11,12,13,14,15,16,17,18,19].
2. Mechanisms of MYB Gene Family as a Transcription Factor
Cellular processes are regulated by transcription factors (TFs), which can modify complex or intricate traits in plants and could play a prominent part in next-generation biotechnology. There are limitations in genomic diversity in traditional breeding. However, transgenic methodologies surpass genetic obstacles by improving the regulatory pathways of one crop by integrating TFs of other crop or plant species [20]. Genes that encode TFs containing DNA binding motifs, e.g., MYB, ERF/AP2, Zinc fingers and bZIP are signal-induced. These TFs further regulate many functional genes during different conditions of stress or morphogenesis. Therefore, identifying novel TF genes responsible for regulating particular gene expression will improve our understating of signaling pathways related to the development and growth of innovative transgenic crops. MYB is a functionally diverse and large protein family present in all eukaryotic organisms [21]. Many MYB acts as TFs having a different number of MYB domains that are able to bind DNA. They interact with other TFs and are also involved in ABA response, which represents their wide distribution among plants. Detailed functional characterization of these proteins in Arabidopsis thaliana depicts their variety of roles in plant-specific mechanisms. The cell cycle of eukaryotes is controlled by ‘classical’ MYB factors that are linked with c-Myb. First, MYB gene identification was form avian virus myeloblastosis, which was ‘oncogene’ v-MYB [22].
3. Evolution of MYB Transcription Factors
The protein of MYB family contains DNA binding domain. There are two particular conserved regions present in MYB protein, C-terminal of R2R3 MYB protein, which show structural and functional diversity in their amino acid sequence, which are responsible for various regulation activities in plants. While, N-terminal show binding domain of MYB DNA are conserved. Generally, the domain of MYB protein comprises sequences with four imperfect amino acid repeats of approximately 52 amino acids, each establishing three α–helices. Ogata, et al. [23] described that Helix-turn-helix structure, which are built through each repeat of second and third helices with regularly spaced three tryptophan residues, resulting in hydrophobic central in HTH structure (3D). Interestingly, first tryptophan in R3 domain is replaced with isoleucine or phenylalanine in plants. MYB family could be separated into four group based on MYB domain number [2,24]. In plants (monocots and dicots), plentiful kind of R2R3-MYB TFs are specific [25]. The plant taxon represents the highest diversity, with the presence of all four classes of MYB proteins. The group of 4R-MYB indicates the smallest class and its members have four R1/R2-type repeats. Several plant genomes contain single 4R-MYB encoded protein. However, the second class retains 3R-MYB protein of R1R2R3 type, which is composed of higher plant genomes, is particularly encoded by five genes. R2R3-MYB domain is more conserved as compared to its other region, which shows more divergence. The division of R2R3-MYB proteins into subgroups is based on amino acid motifs, which are present at C terminal [2].
MYB domain sequence-based evolutionary studies from various organisms represent that plant ancestor initially had three repeats and out of which the first repeat was lost during the course of time. Lipsick [21] has described an evolutionary model of MYB proteins. This model reveals that R1R2R3-MYBs resulted due to consecutive intragenic duplications and triplications among the primeval eukaryotes, and they produced two repeat and three repeat (R1R2R3-MYB, R2R3-MYB) proteins in animals and plants, respectively. During plant evolution through selective subgroup expansion and amplification, numerous subgroups genes harboring R2R3MYB proteins were made due to the loss of R1 [26]. The consecutive gain of repeat units generated MYB genes. The detailed study of MYB genes regarding their classification, structure, characteristics, mechanism of combinational control, multi-functionality, functional redundancy and gain model for evolution have been reviewed comprehensively by Du, et al. [27] and Dubos, Stracke, Grotewold, Weisshaar, Martin and Lepiniec [24]. It is very interesting that the heterogeneous class consists of proteins with partial or single MYB repeat, jointly known as “MYB-related”, which is further divided into many subclasses [28]. The loss of sequence regarding R1 repeat and successive extension of gene family resulted in R2R3-MYB class after evolution from R1R2-MYB gene predecessor [28].
Moreover, it has also been proposed that ancient intragenic duplication by gaining the sequence encoding R1 repeat from R2R3-MYB genes resulted in the evolution of 3R-MYB [29]. Arabidopsis AtMYB48 and AtMYB59 and their rice homologs (OsMYBAS2 and OsMYBAS1), the two R2R3-MYB genes experience alternative splicing in the same way and result in three diverse merged transcripts in rice, and four in Arabidopsis. Therefore, a deep-rooted understanding of another splicing of MYB protein will further enlighten us regarding gene evolution in dicots and monocots, as well as development-related regulation by transcription factor genes [29].
4. Recent Transcriptomic and Genome-Wide Analysis and Expression of MYB Transcription Factors
In sugarcane, 202 MYB TFs are explored, some of them are expressed mainly in stem and are actively responded to drought stress resistance and mosaic diseases [30]. In Arabidopsis 198 MYBs have been identified; among them, 126 are encoded for R2R3-MYB proteins [31]. Recently, 223 MYB (112 R2R3-MYB, 2 R1R2R3-MYB and 119 R1-MYB) transcription factors were recognized in the potato genome [32]. Recently, there are 69 GbMYB transcription factors are identified in Ginkgo biloba, out of which 19 R2R3 MYB are responsive to hormonal and abiotic stresses [33]. In maize, a genome-wide survey indicated that they consist of 157 R2R3-MYB proteins [34]. R2R3-MYB (185) transcription factors are reported in the genome of Mangrove, 34 MYB gene are mainly expressed in different tissues (root, leaves), which are related to various stresses (salinity and drought) [35]. Hippophae rhamnoides is the rich source of secondary metabolites, which has economic importance regarding medicinal and nutritional values, 161 R2R3–MYB TFs were obtained through its genome-wide analysis [36]. In a recent study, 111 StR2R3-MYB transcription factors are reported in potato [37]. In the genome of flax, 167 R2R3-MYB, 7 R3-MYB, and 1R4-MYB transcription factors have been identified [38]. However, in soybean, 252 total MYBs were identified and account for about 4% of all transcription factors. They consist of two (4R-MYB) genes, six (3R-MYB) proteins and 244 encodings for R2R3-MYB proteins [39]. Genome-wide analysis of apples revealed that they contained 229 MYB transcription factors. Another recent study has explored 251 and 305 MYB TFs from Musa balbisiana and Musa acuminata, respectively by [40].
5. Biological Functions Regulated through MYB Transcription Factors
MYB TFs control many plant-specific processes. By using molecular and genetic analysis the function of MYB proteins have been greatly described among various plant species, like petunia (Petunia hybrida), apple (Malus domestica), poplar (Populus tremuloides), snapdragon (Antirrhinum majus), grapevine (Vitis vinifera L.), maize (Zea mays) and Arabidopsis thaliana [41]. R2R3-MYB TFs have been widely investigated during last decade and their involvement in several processes have been revealed, such as abiotic and biotic stress [42,43], cold tolerance [41], phenylpropanoid metabolism [44,45], trichomes development [46], flower shape [47], cell shape [48], plant defense mechanisms [49,50,51,52], cell wall development [53] and stomatal closure [54].
This entry is adapted from the peer-reviewed paper 10.3390/ijms22179544
This entry is offline, you can click here to edit this entry!
