Metabolic engineering is a cutting-edge field that aims to produce simple, readily available, and inexpensive biomolecules by applying different genetic engineering and molecular biology techniques. Fatty acids (FAs) play an important role in determining the physicochemical properties of membrane lipids and are precursors of biofuels. Microbial production of FAs and FA-derived biofuels has several advantages in terms of sustainability and cost. Conventional yeast Saccharomyces cerevisiae is one of the models used for FA synthesis. Several genetic manipulations have been performed to enhance the citrate accumulation and its conversation into acetyl-CoA, a precursor for FA synthesis. Success has been achieved in producing different chemicals, including FAs and their derivatives, through metabolic engineering.
- metabolic engineering technologies
- fatty acid production
- fatty acid in yeast
1. Microbes as Cell Factories in Metabolic Engineering
The sustainable production of biomolecules requires the engineering of microbial cell factories using metabolic engineering principles. They can convert inexpensive feedstocks into chemicals and fuels [1]. The traditional ways to produce these chemicals and fuels have not been able to meet the required demand. Due to economic and ecological goals, microbial production of these chemicals and fuels is growing. However, the efficiency of these microbes to produce these chemicals and fuels is low, which has shifted the focus on engineering microbial-based cells into fully-fledged microbial factories, leading to an alternative approach that has the potential to remove the bottlenecks linked with the other production routes.
Preferably, a microbial cell factory should possess specific characteristics that enable it to produce desired products from the desired feedstock. There are three main features such as (i) enough basic knowledge of the microbe, modeling of media cultures, and bioprocesses, and (ii) the information of dietary requirements. Typically, microbes that can feed on simpler carbon and nitrogen sources are preferred; (iii) resistance to various stresses is crucial since it can affect the overall productivity. Other factors, such as tolerance to harsh conditions, such as high temperature and pH in the fermenter, are also considered [2]. Ideally, the selection of microbial chassis for the biosynthesis of FAs is performed based on their extensive engineering capacity to produce oleochemicals at a high titer [3]. In addition, the engineering strategies in these microorganisms should enhance the supply of metabolic precursors for FAs, such as acetyl-CoA and redox cofactor NADPH.
Moreover, heterogeneous enzymes should also be implemented to functionalize fatty acyl-CoAs into desired derivatives [4]. Currently, model organisms such as E. coli and S. cerevisiae are mostly being utilized as microbial chassis for the synthesis of FAs and are discussed in detail in the reviews [5][6][7]. However, many other promising organisms are also being explored. For instance, Rhodococcus opacus, a chemolithotrophic oleaginous bacterium, has recently been engineered by the deletion of acyl-coenzyme synthetases and the increased expression of three lipase enzymes which had lipase-specific foldase to produce 50.2 gL−1 of Free fatty acids (FFAs) [8].
Robustness is a major requirement while selecting microbes as a cell factory. However, additional challenges are faced during the production of novel compounds using engineered pathways [9]. Classical studies aiming to solve toxicity rely on physiological or cytological effects that lack understanding of the molecular mechanisms of interaction between the microbe and the toxic compounds [10][11]. However, technological advancements have shifted the focus of toxicological assessment to mechanism-centered analysis at the genome-wide level [12]. More recent techniques to deal with toxicity have relied on altering the membrane’s permeability, a concept often called membrane engineering [13]. For instance, the plasma membrane’s oleic acid can be increased through rat elongase 2 gene overexpression, resulting in the increased tolerance of S. cerevisiae to ethanol, propanol, and butanol.
With the development of next-generation DNA assembly and synthesis tools, microbes’ development and characterization as chassis has been in focus. However, not all microbes can be cloned. For instance, more than 99% of the environmental microbes cannot be cultured and studied using the current technologies [14]. Further, foreign genes in a host organism are sometimes expressed weakly or not and are often called “silent” or “cryptic” gene clusters. Other limiting factors include codon optimization between organisms, inadequate precursors and cofactors, and toxicity [15].
2. Genetic Manipulations in FA Production
The majority of the genetic manipulations in microorganisms have targeted E. coli to produce FAs. Plant-based, renewable feedstocks, for example, biomass-derived carbohydrates, have been utilized to produce FAs in E. coli. Steen et al. [16] synthesized fatty esters (biodiesel), fatty alcohols, as well as waxes in engineered E. coli by expressing the native E. coli thioesterase, tesA (a “leaderless” version of TesA), and also eliminated FadD and FadE, the first two competing enzymes which are involved in b-oxidation. Similarly, about 11% more FAs were produced in an engineered E. coli than the control through the over-expression of fabD from four different sources. The well-studied and characterized E. coli fabD gene encodes malonyl CoA-acyl carrier protein transacylase, which catalyzes malonyl-CoA to malonyl-ACP [17]. An increase in the amount of FFAs was observed in the strains having the fabD gene from E. coli, Streptomyces avermitilis MA-4680, or Streptomyces coelicolor A3(2). However, the strain carrying the fabD gene from Clostridium acetobutylicum ATCC 824 was found to produce the same amount of FFAs as the control strain. Notably, the overexpression of the fabD gene was performed by an optimized gene construct. The fabD gene was cloned to form an operon with the TE gene, and hence achieving the overexpression of the gene. The study suggested that FFA production improvements can be brought through the overexpression of the fabD gene to increase malonyl-ACP activity [18].
Similarly, the level of an omega-hydroxy fatty acid, 3-hydroxy propionic acid production, has shown an increased titer up to 1.8 gL−1 h−1, 1.4-fold higher than the wild type. In the study, Chu et al. increased FA production by identifying and cloning novel aldehyde dehydrogenase, GabD4 from Cupriavidus necator. Upon investigation, GabD4_E209Q/E269Q showed the highest enzymatic activity. Metabolic enhancement in E. coli through fatty acyl-ACP reductase from Synechococcus elongatus resulted in improvement of both productivity and yield [19]. The endogenous E. coli AdhP plays a vital role in reducing fatty aldehydes to fatty alcohols, and thus, the study established an encouraging synthetic route for industrial microbial synthesis of fatty acid in method E. coli [20].
Although most of the work for the microbial production of FAs has been performed using E. coli, other organisms have also been utilized. S. cerevisiae has also been a subject of many genetic manipulations for metabolic engineered products, and FAs are not an exception. In yeast, the majority of metabolic fluxes are involved in ethanol production during the fermentation process. Therefore, most enhanced FA production strategies are devised to redirect the fluxes to go through the ethanol pathway. To increase microbial FA production, the main target is to increase the amount of acetyl-CoA, malonyl-CoA, and fatty acyl-CoA. Thus, genetic manipulations for the enhanced synthesis of FAs often target the genes involved in these molecules’ production. Since yeast (S. cerevisiae) does not have enough cytosolic acetyl-CoA production, improving the supply of acetyl-CoA, several genetic manipulations in S. cerevisiae have been conducted. One way to increase the amount of acetyl-CoA production is to inactivate its consumption pathways. The glycolytic flux was redirected towards acetyl-CoA by inactivating the genes for ethanol formation and glycerol production. The inactivation was performed using the widely used marker-based homologous recombination, the loxP–KanMX–loxP method [21]. The inactivation of ADH1, ADH4, GPD1, and GPD2 genes resulted in the production of 100 mg/L FA-derived advanced biofuels, n-butanol [22].
Similarly, in another study, citrate levels were increased by deleting IDH1 and IDH2 genes, which play a role in citrate turnover in the TCA cycle through marker-based homologous recombination. The strategy yielded a 3–4-fold increase in the citrate levels. The excess of citrate was dealt with by the overexpression of a heterologous ATP-citrate lyase (ACL), and hence a significant amount of FAs was recorded in the study [23]. Moreover, Zhou et al. reported 10.4 gL−1 of FFAs, a 20% increase compared to the previously recorded amount, in engineered S. cerevisiae by optimizing a synthetic citrate lyase pathway. This was done by expressing the ATP citrate lyase and malic enzyme [24]. Alongside the optimization of the two genes, the upregulation of the native mitochondrial citrate transporter and malate dehydrogenase was also performed in the study. Additionally, the malate synthesis pathway was tweaked to increase the production of FAs. For instance, FA production was increased from 460 to 780 mg/L, a 70% increase [25], by downregulating malate synthase, cloning of ATP citrate lyase from Y. lipolytica, and eliminating the expression of cytoplasmic glycerol-3-phosphate dehydrogenase. Altogether, due to these genetic manipulations, 0.8 g/L of FFAs in shake flasks was recorded.
Malonyl-CoA, another important precursor for the biosynthesis of FAs in S. cerevisiae, is catalyzed by acetyl-CoA carboxylase (ACC) from acetyl-CoA. Increasing the supply of malonyl-CoA is vital for increasing FAs in yeast. The increased expression of the ACC1 gene (encodes ACC) and FAS gene yields a higher level of malonyl-CoA. In S. cerevisiae, the overexpression of ACC1, FAS1, and FAS2 enhanced the titer of FAs [26]. Moreover, heterogeneous expression of genes to obtain an increased supply of malonyl-CoA has also been performed. For instance, FAS expression from Brevibacterium ammoniagenes in S. cerevisiae resulted in producing 10,498 μg FAEE/g CDW of fatty acid ethyl ester, an increase of about 6.2-fold compared to the wild type [27]. In another study, a more direct approach to increasing the supply of malonyl-CoA was taken. AAE13, a plant malonyl-CoA synthetase gene overexpression in S. cerevisiae, led to the increased accumulation of lipid and resveratrol of 1.6-times and 2.4-times, respectively [28]. More recently, dCas9 and malonyl-CoA responsive intracellular biosensors were used to identify novel gene expression fine-tuning set-ups to enhance the levels of acetyl-CoA and malonyl-CoA. A total of 3194 gRNAs were used to target 168 selected genes, followed by screening potential malonyl-CoA overproducers using fluorescence-activated sorting assay [29].
On the other hand, another way to increase the oleaginous microbes’ efficiency by increasing NADPH supply is to reduce the acetyl groups (CH3–CO–) in the growing acyl chain of FAs (–CH2– CH2–). In most oleaginous species, the malic enzyme provides most of the NADPH for FA biosynthesis [30]. Additionally, the pentose phosphate pathway (PPP) provides the additional NADPH required. In order to increase the production of FAs, the enzyme involved has been manipulated. A study by Hao et al. [31] shows the overexpression of the genes for glucose-6-phosphate dehydrogenase (G6PD) and 6-phosphogluconate dehydrogenase (6PGD), the enzymes responsible for increasing the supply of NADPH. Because of the overexpression of the genes, the engineered strain produced 8.1 ± 0.5 g/L FAs, which is an increase of 1.7-times [31].
3. Yeast and Production of FAs Using Metabolic Engineering
Yeast is an important organism that has been used extensively as a model organism for molecular biology studies. It is the first eukaryotic organism whose genome has been sequenced [32]. Compared to molds and algae and tolerance towards harsh conditions in industrial production, its high growth rate contributes to the popularity of oleaginous yeast as an ideal organism for producing FAs and next-generation biofuels [33].
A large breadth of genetic information is available about the yeast genome. Various specific databases are available, containing genetics, proteomics, and interactomics information (Table 1). The information has been handy in considering which genes should be selected as a target for metabolic enhancement. Winzeler et al., in 1999, performed splendid work on a project popularly called The Saccharomyces Genome Deletion Project [34]. During the project, 6925 Saccharomyces cerevisiae strains were constructed, knocking out almost 96% of the yeast genome. The project provided unique knowledge of over 6000 gene disruption mutants and proved to be of great importance for future metabolic engineers. Apart from metabolic engineering uses, yeast has also become a reliable model organism for studying various fundamental studies such as the cell cycle [35], numerous cancers [36], and viruses [37]. Generally, there are two methods of designing yeast strains: one involves the introduction of foreign genes to modify its metabolic pathways that can face regulatory issues, and the other without inducing any DNA edits or only introducing genetic material limited to the genus Saccharomyces, which is often called self-cloning [38]. Since these genetic perturbations may also occur naturally by breeding and self-cloning that’s why in several countries, they are acceptable.
Table 1. Yeast-related online databases and their web links.
| Database | Website |
|---|---|
| Saccharomyces Genome database | http://genome-www.stanford.edu/ (accessed on 25 June 2021) |
| Yeast deletion project | http://www-sequence.stanford.edu/group/yeast_deletion_project/deletions3.html (accessed on 25 June 2021) |
| Transcriptional regulatory code of yeast | http://younglab.wi.mit.edu/regulatory_code/ (accessed on 25 June 2021) |
| Yeast GFP fusion localization database | https://yeastgfp.yeastgenome.org/ (accessed on 25 June 2021) |
| General repository of interaction datasheets | https://thebiogrid.org/ (accessed on 25 June 2021) |
| Yeast search for transcriptional regulators and consensus tracking | http://yeastract.com/ (accessed on 25 June 2021) |
| Cold Spring Harbor laboratory | https://reactome.org/ (accessed on 25 June 2021) |
In yeast, FA biosynthesis starts with converting acetyl-CoA to malonyl-CoA through the enzyme acetyl-CoA carboxylase in the cytosol. The precursors, malonyl-CoA, and acetyl-CoA are condensed by FA synthases (FASs) to FAs using malonyl-CoA as the extender unit. Each elongation of two-carbon units in FA biosynthesis requires two NADPH. FAs in the cytosol of S. cerevisiae are catalyzed by the type I FAS system and have two functional subunits: the α-subunit and β-subunit encoded by FAS2 and FAS1, respectively [39][40]. Although type II FAS is also responsible for FAs, most FAs in S. cerevisiae are synthesized by the type I FAS system. Principally, there are two ways of generating acetyl-CoA in yeast cells. First, CoA is generated from the glycolysis of fermentable sugars through the pyruvate–acetaldehyde–acetate pathway through cytoplasmic acetyl-CoA synthases activity. The second source for acetyl-CoA synthesis in S. cerevisiae is the excess of citrate (Figure 1). This excess amount of citrate is transported with the help of citrate transport protein [41]. Citrate is finally catalyzed by ATP-citrate lyase, an enzyme present in all oil-producing microorganisms [42].
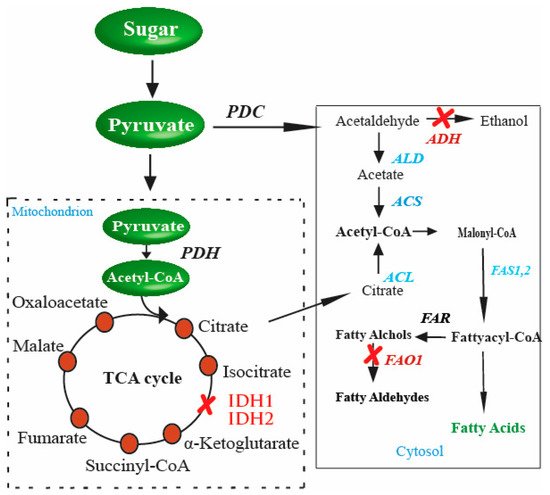
Figure 1. Genetic perturbations in S. cerevisiae for enhanced production of FAs. These genetic perturbations result in higher levels of FAs in S. cerevisiae. Once there is an excess of citrate in the cytosol, the gene responsible for converting citrate to acetyl-CoA, ACL (shown in red in the cytosol), is inserted into S. cerevisiae. These genetic perturbations result in higher levels of FAs in yeast. The strengthened steps involved in enhanced FA production are indicated in blue, and the blocked steps are indicated in red color.
The possible way for the metabolic improvement of acetyl-CoA production, as proposed by [43], is first to increase the amount of citrate accumulated in the tricarboxylic acid cycle (TCA cycle), and then transport the excess of citrate in order to generate cytosol acetyl-CoA. This is a promising pathway to provide precursors for FA biosynthesis. Another aspect of the improved production of FAs in S. cerevisiae is citrate catabolism. During the TCA cycle, citrate catabolism is performed by an enzyme of mitochondrial origin, isocitrate dehydrogenase. The enzyme is coded by two distinct genes, IDH1 and IDH2 [44]. The accumulation of citrate is directly linked to its consumption and hence to the enzyme activity, i.e., if the enzyme activity is high, there is less accumulation of citrate and less chance of producing a high amount of acetyl-CoA. To achieve high production, the enzyme activity needs to be reduced. This is performed by disturbing the genes IDH1 and IDH2 separately, and then the effect on citrate accumulation is measured compared to the wild-type strains. The double gene disturbance resultantly yielded a 4 to 5-fold enhancement in citrate level in the media. This suggested that since two genes and those genes code, the enzyme that catalyzes citrate was disturbed in the process, resulting in the net increase in citrate accumulation in the media [23].
Furthermore, the production of a surplus amount of citrate is not only required; instead, the accumulated citrate should also be converted into cytosolic acetyl-CoA. This is performed by the enzyme ATP-citrate lyase (ACL). Several studies regarding the effect of ACL on the cytoplasmic acetyl-CoA generation or FA accumulation have been conducted [45][46]. These studies established a relationship between ACL action and acetyl-CoA precursor synthesis for FA production. However, despite being present in various organisms, including human cells, animal cells, plant cells, and in certain species of yeasts, the enzyme is absent in the Saccharomycotina. Therefore, an obvious target for improving acetyl-CoA production and eventually increasing FA synthesis is the insertion of genes that code for the enzymes responsible for catalyzing the accumulated citrate to acetyl-CoA in the cytosol [23]. Similarly, in another study, pyruvate was directed towards endogenous cytosolic acetyl-CoA biosynthesis by deleting IDH1 and expressing ACL from Aspergillus nidulans for production mevalonate [47]. This strategy of directing flux through the ACL pathway by increasing citrated supply and the expression of active ACL can be applied to synthesize acetyl-CoA-derived molecules such as FAs.
This entry is adapted from the peer-reviewed paper 10.3390/biology10070632
References
- Keasling, J.D. Manufacturing molecules through metabolic engineering. Science 2010, 330, 1355–1358.
- Calero, P.; Nikel, P.I. Chasing bacterial chassis for metabolic engineering: A perspective review from classical to non-traditional microorganisms. Microb. Biotechnol. 2019, 12, 98–124.
- Jin, M.; Slininger, P.J.; Dien, B.S.; Waghmode, S.; Moser, B.R.; Orjuela, A.; Sousa, L.d.C.; Balan, V. Microbial lipid-based lignocellulosic biorefinery: Feasibility and challenges. Trends Biotechnol. 2015, 33, 43–54.
- Marella, E.R.; Holkenbrink, C.; Siewers, V.; Borodina, I. Engineering microbial fatty acid metabolism for biofuels and biochemicals. Curr. Opin. Biotechnol. 2018, 50, 39–46.
- Yang, D.; Park, S.Y.; Park, Y.S.; Eun, H.; Lee, S.Y. Metabolic Engineering of Escherichia coli for Natural Product Biosynthesis. Trends Biotechnol. 2020, 38, 745–765.
- Hu, Y.; Zhu, Z.; Nielsen, J.; Siewers, V. Engineering Saccharomyces cerevisiae cells for production of fatty acid-derived biofuels and chemicals. Open Biol. 2019.
- Kim, S.K.; Park, Y.C. Biosynthesis of ω-hydroxy fatty acids and related chemicals from natural fatty acids by recombinant Escherichia coli. Appl. Microbiol. Biotechnol. 2019.
- Kim, H.M.; Chae, T.U.; Choi, S.Y.; Kim, W.J.; Lee, S.Y. Engineering of an oleaginous bacterium for the production of fatty acids and fuels. Nat. Chem. Biol. 2019.
- Nicolaou, S.A.; Gaida, S.M.; Papoutsakis, E.T. A comparative view of metabolite and substrate stress and tolerance in microbial bioprocessing: From biofuels and chemicals, to biocatalysis and bioremediation. Metab. Eng. 2010.
- Pinkart, H.C.; White, D.C. Phospholipid biosynthesis and solvent tolerance in Pseudomonas putida strains. J. Bacteriol. 1997.
- Sikkema, J.; De Bont, J.A.M.; Poolman, B. Mechanisms of membrane toxicity of hydrocarbons. Microbiol. Rev. 1995, 59, 201–222.
- Dos Santos, S.C.; Teixeira, M.C.; Cabrito, T.R.; Sá-Correia, I. Yeast toxicogenomics: Genome-wide responses to chemical stresses with impact in environmental health, pharmacology, and biotechnology. Front. Genet. 2012.
- Dunlop, M.J. Engineering microbes for tolerance to next-generation biofuels. Biotechnol. Biofuels 2011.
- Daniel, R. The metagenomics of soil. Nat. Rev. Microbiol. 2005.
- Luo, Y.; Enghiad, B.; Zhao, H. New tools for reconstruction and heterologous expression of natural product biosynthetic gene clusters. Nat. Prod. Rep. 2016.
- Steen, E.J.; Kang, Y.; Bokinsky, G.; Hu, Z.; Schirmer, A.; McClure, A.; Del Cardayre, S.B.; Keasling, J.D. Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature 2010.
- Joshi, V.C. Mechanism of malonyl-coenzyme A-acyl-carrier protein transacylase. Biochem. J. 1972.
- Zhang, X.; Agrawal, A.; San, K.Y. Improving fatty acid production in escherichia coli through the overexpression of malonyl coA-Acyl carrier protein transacylase. Biotechnol. Prog. 2012.
- Chu, H.S.; Kim, Y.S.; Lee, C.M.; Lee, J.H.; Jung, W.S.; Ahn, J.H.; Song, S.H.; Choi, I.S.; Cho, K.M. Metabolic engineering of 3-hydroxypropionic acid biosynthesis in Escherichia coli. Biotechnol. Bioeng. 2015.
- Liu, R.; Zhu, F.; Lu, L.; Fu, A.; Lu, J.; Deng, Z.; Liu, T. Metabolic engineering of fatty acyl-ACP reductase-dependent pathway to improve fatty alcohol production in Escherichia coli. Metab. Eng. 2014.
- Hegemann, J.H.; Gldener, U.; Köhler, G.J. Gene disruption in the budding yeast Saccharomyces cerevisiae. Methods Mol. Biol. 2006.
- Lian, J.; Si, T.; Nair, N.U.; Zhao, H. Design and construction of acetyl-CoA overproducing Saccharomyces cerevisiae strains. In Proceedings of the Food, Pharmaceutical and Bioengineering Division 2014—Core Programming Area at the 2014 AIChE Annual Meeting, Atlanta, GA, USA, 16–21 November 2014.
- Tang, X.; Feng, H.; Chen, W.N. Metabolic engineering for enhanced fatty acids synthesis in Saccharomyces cerevisiae. Metab. Eng. 2013.
- Zhou, Y.J.; Buijs, N.A.; Zhu, Z.; Qin, J.; Siewers, V.; Nielsen, J. Production of fatty acid-derived oleochemicals and biofuels by synthetic yeast cell factories. Nat. Commun. 2016.
- Ghosh, A.; Ando, D.; Gin, J.; Runguphan, W.; Denby, C.; Wang, G.; Baidoo, E.E.K.; Shymansky, C.; Keasling, J.D.; Martín, H.G. 13C metabolic flux analysis for systematic metabolic engineering of S. cerevisiae for overproduction of fatty acids. Front. Bioeng. Biotechnol. 2016.
- Runguphan, W.; Keasling, J.D. Metabolic engineering of Saccharomyces cerevisiae for production of fatty acid-derived biofuels and chemicals. Metab. Eng. 2014.
- Eriksen, D.T.; HamediRad, M.; Yuan, Y.; Zhao, H. Orthogonal Fatty Acid Biosynthetic Pathway Improves Fatty Acid Ethyl Ester Production in Saccharomyces cerevisiae. ACS Synth. Biol. 2015.
- Wang, Y.; Chen, H.; Yu, O. A plant malonyl-CoA synthetase enhances lipid content and polyketide yield in yeast cells. Appl. Microbiol. Biotechnol. 2014.
- Ferreira, R.; Skrekas, C.; Hedin, A.; Sánchez, B.J.; Siewers, V.; Nielsen, J.; David, F. Model-Assisted Fine-Tuning of Central Carbon Metabolism in Yeast through dCas9-Based Regulation. ACS Synth. Biol. 2019.
- Ratledge, C.; Wynn, J.P. The biochemistry and molecular biology of lipid accumulation in oleaginous microorganisms. Adv. Appl. Microbiol. 2002, 51, 1–52.
- Hao, G.; Chen, H.; Gu, Z.; Zhang, H.; Chen, W.; Chen, Y.Q. Metabolic engineering of Mortierella alpina for enhanced arachidonic acid production through the NADPH-supplying strategy. Appl. Environ. Microbiol. 2016.
- Goffeau, A.; Barrell, G.; Bussey, H.; Davis, R.W.; Dujon, B.; Feldmann, H.; Galibert, F.; Hoheisel, J.D.; Jacq, C.; Johnston, M.; et al. Life with 6000 genes. Science 1996.
- Li, Y.; Zhao, Z.; Bai, F. High-density cultivation of oleaginous yeast Rhodosporidium toruloides Y4 in fed-batch culture. Enzyme Microb. Technol. 2007.
- Winzeler, E.A.; Shoemaker, D.D.; Astromoff, A.; Liang, H.; Anderson, K.; Andre, B.; Bangham, R.; Benito, R.; Boeke, J.D.; Bussey, H.; et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 1999.
- Breeden, L.L. Periodic transcription: A cycle within a cycle. Curr. Biol. 2003.
- Spradling, A.; Ganetsky, B.; Hieter, P.; Johnston, M.; Olson, M.; Orr-Weaver, T.; Rossant, J.; Sanchez, A.; Waterston, R. New roles for model genetic organisms in understanding and treating human disease: Report from the 2006 Genetics Society of America meeting. Genetics 2006.
- Galao, R.P.; Scheller, N.; Alves-Rodrigues, I.; Breinig, T.; Meyerhans, A.; Díez, J. Saccharomyces cerevisiae: A versatile eukaryotic system in virology. Microb. Cell Fact. 2007.
- Kitagaki, H.; Kitamoto, K. Breeding research on sake yeasts in Japan: History, recent technological advances, and future perspectives. Annu. Rev. Food Sci. Technol. 2013.
- Hiltunen, J.K.; Schonauer, M.S.; Autio, K.J.; Mittelmeier, T.M.; Kastaniotis, A.J.; Dieckmann, C.L. Mitochondrial fatty acid synthesis type II: More than just fatty acids. J. Biol. Chem. 2009.
- Chan, D.I.; Vogel, H.J. Current understanding of fatty acid biosynthesis and the acyl carrier protein. Biochem. J. 2010.
- Anoop, V.M. Modulation of Citrate Metabolism Alters Aluminum Tolerance in Yeast and Transgenic Canola Overexpressing a Mitochondrial Citrate Synthase. Plant Physiol. 2003.
- Hynes, M.J.; Murray, S.L. ATP-Citrate Lyase Is Required for Production of Cytosolic Acetyl Coenzyme A and Development in Aspergillus nidulans. Eukaryot. Cell 2010.
- Ratledge, C.; Bowater, M.D.V.; Taylor, P.N. Correlation of ATP/citrate lyase activity with lipid accumulation in developing seeds of Brassica napus L. Lipids 1997.
- Wang, J.; Jiang, J.C.; Jazwinski, S.M. Gene regulatory changes in yeast during life extension by nutrient limitation. Exp. Gerontol. 2010.
- Wellen, K.E.; Hatzivassiliou, G.; Sachdeva, U.M.; Bui, T.V.; Cross, J.R.; Thompson, C.B. ATP-citrate lyase links cellular metabolism to histone acetylation. Science 2009.
- Rangasamy, D.; Ratledge, C. Genetic Enhancement of Fatty Acid Synthesis by Targeting Rat Liver ATP:Citrate Lyase into Plastids of Tobacco. Plant Physiol. 2002.
- Rodriguez, S.; Denby, C.M.; Vu, T.; Baidoo, E.E.K.; Wang, G.; Keasling, J.D. ATP citrate lyase mediated cytosolic acetyl-CoA biosynthesis increases mevalonate production in Saccharomyces cerevisiae. Microb. Cell Fact. 2016.
