Choriocarcinoma (CC), a subtype of trophoblastic disease, is a rare and highly aggressive neoplasm. There are two main CC subtypes: gestational and non-gestational, (so called when it develops as a component of a germ cell tumor or is related to a somatic mutation of a poorly differentiated carcinoma), each with very diverse biological activity. Long non-coding (lnc) RNAs are non-coding transcripts that are longer than 200 nucleotides. LncRNAs can act as oncogenes or tumor suppressor genes. Deregulation of their expression has a key role in tumor development, angiogenesis, differentiation, migration, apoptosis, and proliferation.
- choriocarcinoma
- rare cancer
- long non-coding RNA
- oncogenes or tumor suppressor genes
- lncRNA-based therapy
1. Introduction
| Types | Gestational Choriocarcinoma | Non-Gestational Choriocarcinoma | |
|---|---|---|---|
| Germ Cell Tumor | Somatic Carcinoma | ||
| Incidence | Ranges from 1 in Europe to −9.2 in Asia/40,000 pregnancies | Rare < 1% of all ovarian tumors—children, young adults but rarely in older adults. Midline tumors mostly in males | Rare ovarian carcinomas in adults |
| Origin | It may develop as a complication of pregnancy, usually following a complete mole | It arises from primordial germ cells | It arises from differentiation of pluripotent cells into a somatic carcinoma |
| Site | Primarily uterus and also intraplacental; rarely ovary and extrauterine sites | Gonads, midline: pineal gland, mediastinum, retroperitonum | Lung, gastrointestinal tract, and other organs, including very rare ovarian carcinoma and uterine cases in post-menopause |
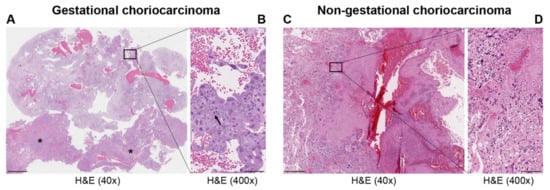
| Histopathology | Mononuclear cytotrophoblast and intermediate trophoblast and multinucleated syncytiotrophoblast cells with marked atypia and mitoses | Mainly in pure form with cyto- and syncytiotrophoblast or with other components of germ cell tumors (mixed germ cell tumor) | Presence hCC-producing multinucleated giant cells; transition with co-existing somatic carcinoma of the particular organ |
| Cytogenetic features | Deletion of 7p12-7q11.2; amplification of 7q21-q31 and loss of 8p12-21 [3] | Gain of 12p [3] | Unknown |
| Biochemical features | hCG in serum or urine (>10 × 103 mlU/mL) | hCG in serum or urine | hCG in serum or urine—variable |
| Molecular markers | Upregulation of TP53, CDKN1A, RB1, EGFR, ERBB2, c-MYC, BCL2, NANOG, H19 [3][8]; Downregulation of NECC1, TIMP3, DOC-2/hDab2, RASSF1A, CDKN2A, CDH1, IGF2, OCT4, SOX2 [3][8]; Mutated genes: NLRP7, ARID1A, SMARCD1, EP300 [9] |
Upregulation of CGB5, CGA, NANOG, STELLA, GDF3 [3] | Upregulation of NANOG [3] |
| Treatment | Chemotherapy | Surgery is indicated. Chemotherapy of different drug regimens is applied | Surgery is indicated. May respond to chemotherapy but it may not be useful |
| Prognosis | Good | Poor | Poor |
2. Dysregulated lncRNAs in Choriocarcinoma
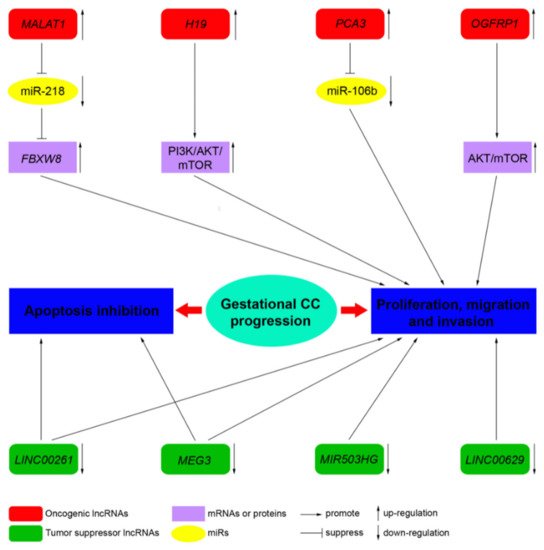
LncRNAs, by acting as tumor suppressor genes or oncogenes, play a critical physiological role in apoptosis, invasion, metastasis, and cell proliferation in several cancers. LncRNAs are also involved in the pathogenesis of cancers of the female reproductive system (ovarian, uterine, vaginal, cervical, and vulvar cancers) [10]. However, to date, only a few studies address the role of lncRNA in CC. Novel mechanistic insights into how gene expression is specifically regulated by lncRNAs, contributing to CC formation, are outlined below. Table 2 summarizes the functions of the main lncRNAs implicated in CC.
| LncRNA | Locus | Role | Molecular Functions | Target Pathway | Sources |
|---|---|---|---|---|---|
| MALAT1 | 11q13.1 | Oncogene | Sponge of miR-218 | Unknown | Three CC cell lines, JEG-3, JAR, and BeWo cells, and a normal cell line human trophoblast cells (HT cells) [11] |
| H19 | 11p15.5 | Oncogene | Unknown | PI3K/AKT/mTOR [12] | Placenta, androgenetic moles, and choriocarcinoma [13]; CC cell line JEG-3, including MTX- and 5-FU-resistant variants [12] |
| MEG3 | 14q32.3 | Tumor suppressor | Unknown | Unknown | Placenta; 4 cell-lines associated with pregnancy, including HTR-8/SVneo, JEG-3, WISH, and HUVEC [14] |
| PCA3 | 9q21-22 | Oncogene | Sponge of miR-106b | Unknown | Three CC cell lines, JAR, BeWo, and JEG-3, and the human chorionic trophoblast cell HTR-8 [15] |
| LINC00261 | 20p11.21 | Tumor suppressor | Unknown | Unknown | Sixty CC tissues and 60 adjacent non-cancerous tissues; 3 CC cell lines, namely, BeWo CCL-98, JEG-3, and JAR [16] |
| OGFRP1 | 22q13.2 | Oncogene | Unknown | AKT/mTOR | Two CC cell lines, JEG-3 and JAR [17] |
| MIR503HG and LINC00629 | Xq26 | Tumor suppressor | Unknown | Unknown | RNA samples from a commercial normal human tissue panel and 18 cancer cell lines, JEG-3 cell line [18] |
3. Clinical Applications of lncRNAs in Choriocarcinoma
Given that lncRNAs can be detected in almost all tissues and body fluids, including peripheral blood, and are not easily degraded by RNases, they can be more sensitive and specific than DNA, proteins, and protein-coding RNAs in the diagnosis of tumors [19][20]. Thus, lncRNAs can be potentially utilized as novel non-invasive biomarkers in cancer diagnosis [21]. Even though there has been increasing interest in the study of lncRNAs in gynecological cancers, research is still preliminary. The association of many dysregulated lncRNAs with clinical features leads to the design of novel biomarkers for diagnosis and management of patients with gynecological cancers, ultimately attaining better prognosis [10]. Whether lncRNAs will be also beneficial in the early detection of CC needs to be investigated. Currently, there is no evidence that lncRNAs are used as biomarkers for diagnosis and prognosis of patients with CC. Moreover, given the rarity of the tumor, we hope that these molecules may be used in the future to distinguish gestational CC from non-gestational tumors in an easier and quicker way than the current state-of-the-art methods (biopsy and microsatellite analysis).
4. LncRNAs as Therapeutic Targets
Interestingly, most lncRNAs are expressed in a cell type- and tissue-specific manner, and in this respect, lncRNAs lend themselves as potentially important therapeutic targets. Specifically, lncRNA targeted therapy, unlike chemotherapeutic regimens, needs to hinder the signaling pathways in which lncRNA plays an important role in tumor development and progression, whilst aiming to avoid the adverse effects on normal cells. There may also be a potential role of lncRNA as biomarkers for disease management, including non-invasive screening [22].
Recent advances utilize CRISPR technology and oligonucleotide-based therapy to modify gene expression. These approaches may be pivotal for developing novel therapeutic approaches aiming at interfering with oncogenic lncRNAs [23].
The use of RNA interference (RNAi) to modulate gene expression has been successfully used to silence lncRNA in vivo [24]. Advancements in RNAi have led to Food and Drug Administration (FDA) approval of lipid nanoparticles (LNPs) which contain short interfering RNA (siRNA) that is being used to treat hereditary amyloidosis in adults [25].
LncRNAs associated with cancer can also be modulated by using antisense oligonucleotides (ASOs). ASOs are single stranded with a central native or chemically modified DNA stretch, flanked on either side by RNA nucleotides. DNA forms RNA/DNA heteroduplex with target lncRNA, which is cleaved by endogenous RNaseH1 [26]. ASOs have been successfully used to alter mRNA expression as part of the treatment of various diseases, including different types of cancer where lncRNAs are highly expressed [27][28].
Advances in genomic interference methods, with superior specificity when compared to RNAi, have been developed recently. CRISPR interference (CRISPRi) using CRISPR-Cas9 and CRISPR-Cas13 led to the successful silencing of transcriptionally active lncRNA-expressing sites. In the CRISPR-Cas9 approach, nucleotides devoid of nucleolytic activity, termed dead-Cas9, are fused to transcriptional repressors. Transcriptional silencing is achieved because, via guide RNAs, this fusion protein is targeted to a specific gene promoter [29]. The development of guide RNAs targeting the promoters of thousands of lncRNAs in the human genome together with CRISPRi enabled the selective inactivation of lncRNA genes in human cancer cell lines. Liu et al. further underscored tissue specificity of lncRNA by identifying approximately 500 lncRNAs in only one cell type [30]. Therefore, it is possible to achieve transcriptional silencing of lncRNAs via CRISPR-based approaches, however, several challenges limit their immediate use for therapeutic targeting [31][32][33][34]. Major limitations include delivery methods (viral and non-viral vectors), with viral vectors limited in the size of the cargo they are able to deliver to the cells of interest [35][36][37][38]. Regardless of the current limitations, the future undoubtedly looks bright for CRISPR-directed lncRNA therapies.
5. Future Directions and Conclusions
This entry is adapted from the peer-reviewed paper 10.3390/ijms22126506
References
- Bruce, S.; Sorosky, J. Gestational Trophoblastic Disease. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2021. Available online: (accessed on 18 February 2021).
- Lurain, J.R. Gestational trophoblastic disease I: Epidemiology, pathology, clinical presentation and diagnosis of gestational trophoblastic disease, and management of hydatidiform mole. Am. J. Obstet. Gynecol. 2010, 203, 531–539.
- Cheung, A.N.; Zhang, H.J.; Xue, W.C.; Siu, M.K. Pathogenesis of choriocarcinoma: Clinical, genetic and stem cell perspectives. Future Oncol. 2009, 5, 217–231.
- Stockton, L.; Green, E.; Kaur, B.; de Winton, E. Non-Gestational Choriocarcinoma with Widespread Metastases Presenting with Type 1 Respiratory Failure in a 39-Year-Old Female: Case Report and Review of the Literature. Case Rep. Oncol. 2018, 11, 151–158.
- WHO. Classification of tumors editoral board. Female genital tumors. In WHO Classification of Tumors Series, 5th ed.; IARC: Lyon, France, 2020; Volume 4, Available online: (accessed on 20 December 2020).
- Zhang, X.; Yan, K.; Chen, J.; Xie, X. Using short tandem repeat analysis for choriocarcinoma diagnosis: A case series. Diagn. Pathol. 2019, 14, 93.
- Friedlander, T.W.; Ryan, C.J.; Small, E.J.; Torti, F. Testicular cancer. In Abeloff’s Clinical Oncology, 5th ed.; Niederhuber, J.E., Armitage, J.O., Doroshow, J.H., Kastan, M.B., Tepper, J.E., Eds.; Elsevier Saunders: Philadelphia, PA, USA, 2014; Chapter 86; pp. 1507–1533.
- Shih, I.-M. Gestational trophoblastic neoplasia—Pathogenesis and potential therapeutic targets. Lancet Oncol. 2007, 8, 642–650.
- Jung, S.H.; Choi, Y.J.; Kim, M.S.; Park, H.C.; Han, M.R.; Hur, S.Y.; Lee, A.W.; Shin, O.R.; Kim, J.; Lee, S.H. Distinct genomic profiles of gestational choriocarcinoma, a unique cancer of pregnant tissues. Exp. Mol. Med. 2020, 52, 2046–2054.
- Hosseini, E.S.; Meryet-Figuiere, M.; Sabzalipoor, H.; Kashani, H.H.; Nikzad, H.; Asemi, Z. Dysregulated expression of long noncoding RNAs in gynecologic cancers. Mol. Cancer 2017, 16, 107.
- Shi, D.; Zhang, Y.; Lu, R.; Zhang, Y. The long non-coding RNA MALAT1 interacted with miR-218 modulates choriocarcinoma growth by targeting Fbxw8. Biomed. Pharmacother. 2018, 97, 543–550.
- Yu, S.; Wu, C.; Tan, Q.; Liu, H. Long noncoding RNA H19 promotes chemotherapy resistance in choriocarcinoma cells. J. Cell. Biochem. 2019, 120, 15131–15144.
- Wake, N.; Arima, T.; Matsuda, T. Involvement of IGF2 and H19 imprinting in choriocarcinoma development. Int. J. Gynaecol. Obstet. 1998, 60, S1–S8.
- Zhang, Y.; Zou, Y.; Wang, W.; Zuo, Q.; Jiang, Z.; Sun, M.; De, W.; Sun, L. Down-regulated long non-coding RNA MEG3 and its effect on promoting apoptosis and suppressing migration of trophoblast cells. J. Cell. Biochem. 2015, 116, 542–550.
- Wang, Y.N.; Liu, S.Y.; Wang, L.; Han, L.Y. Long noncoding RNA PCA3 contributes to the progression of choriocarcinoma by acting as a ceRNA against miR-106b. Int. J. Clin. Exp. Pathol. 2019, 12, 1609–1617.
- Wang, Y.; Xue, K.; Guan, Y.; Jin, Y.; Liu, S.; Wang, Y.; Liu, S.; Wang, L.; Han, L. Long Noncoding RNA LINC00261 Suppresses Cell Proliferation and Invasion and Promotes Cell Apoptosis in Human Choriocarcinoma. Oncol. Res. 2017, 25, 733–742.
- Meng, Q.; Xue, H. Knockdown of lncRNA OGFRP1 Inhibits Proliferation and Invasion of JEG-3 Cells Via AKT/mTOR Pathway. Technol. Cancer Res. Treat. 2020, 19, 1533033820905823.
- Muys, B.R.; Lorenzi, J.C.; Zanette, D.L.; Lima e Bueno Rde, B.; de Araújo, L.F.; Dinarte-Santos, A.R.; Alves, C.P.; Ramão, A.; de Molfetta, G.A.; Vidal, D.O.; et al. Placenta-Enriched LincRNAs MIR503HG and LINC00629 Decrease Migration and Invasion Potential of JEG-3 Cell Line. PLoS ONE 2016, 11, e0151560.
- Xu, M.D.; Qi, P.; Du, X. Long non-coding RNAs in colorectal cancer: Implications for pathogenesis and clinical application. Mod. Pathol. 2014, 27, 1310–1320.
- Traboulsi, W.; Sergent, F.; Boufettal, H.; Brouillet, S.; Slim, R.; Hoffmann, P.; Benlahfid, M.; Zhou, Q.Y.; Balboni, G.; Onnis, V. Antagonism of EG-VEGF Receptors as Targeted Therapy for Choriocarcinoma Progression In Vitro and In Vivo. Clin. Cancer Res. 2017, 23, 7130–7140.
- Kishikawa, T.; Otsuka, M.; Ohno, M.; Yoshikawa, T.; Takata, A.; Koike, K. Circulating RNAs as new biomarkers for detecting pancreatic cancer. World J. Gastroenterol. 2015, 21, 8527–8540.
- Arun, G.; Diermeier, S.D.; Spector, D.L. Therapeutic Targeting of Long Non-Coding RNAs in Cancer. Trends Mol. Med. 2018, 24, 257–277.
- Schwarzmueller, L.; Bril, O.; Vermeulen, L.; Léveillé, N. Emerging Role and Therapeutic Potential of lncRNAs in Colorectal Cancer. Cancers 2020, 12, 3843.
- Borel, F.; Kay, M.A.; Mueller, C. Recombinant AAV as a platform for translating the therapeutic potential of RNA interference. Mol. Ther. 2014, 22, 692–701.
- Adams, D.; Gonzalez-Duarte, A.; O’Riordan, W.D.; Yang, C.C.; Ueda, M.; Kristen, A.V.; Tournev, I.; Schmidt, H.H.; Coelho, T.; Berk, J.L.; et al. Patisiran, an RNAi Therapeutic, for Hereditary Transthyretin Amyloidosis. N. Engl. J. Med. 2018, 379, 11–21.
- Walder, R.Y.; Walder, J.A. Role of RNase H in hybrid-arrested translation by antisense oligonucleotides. Proc. Natl. Acad. Sci. USA 1988, 85, 5011–5015.
- Chiriboga, C.A. Nusinersen for the treatment of spinal muscular atrophy. Expert Rev. Neurother. 2017, 17, 955–962.
- Jaschinski, F.; Rothhammer, T.; Jachimczak, P.; Seitz, C.; Schneider, A.; Schlingensiepen, K.H. The antisense oligonucleotide trabedersen (AP12009) for the targeted inhibition of TGF-beta2. Curr. Pharm. Biotechnol. 2011, 12, 2203–2213.
- Brezgin, S.; Kostyusheva, A.; Kostyushev, D.; Chulanov, V. Dead Cas Systems: Types, Principles, and Applications. Int. J. Mol. Sci. 2019, 20, 6041.
- Liu, S.J.; Horlbeck, M.A.; Cho, S.W. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science 2017, 355.
- Ates, I.; Rathbone, T.; Stuart, C.; Bridges, P.H.; Cottle, R.N. Delivery approaches for therapeutic genome editing and challenges. Genes 2020, 11, 1113.
- Lino, C.A.; Harper, J.C.; Carney, J.P.; Timlin, J.A. Delivering crispr: A review of the challenges and approaches. Drug Deliv. 2018, 25, 1234–1257.
- Witzigmann, D.; Kulkarni, J.A.; Leung, J.; Chen, S.; Cullis, P.R.; van der Meel, R. Lipid nanoparticle technology for therapeutic gene regulation in the liver. Adv. Drug Deliv. Rev. 2020, 159, 344–363.
- Abudayyeh, O.O.; Gootenberg, J.S.; Essletzbichler, P.; Han, S.; Joung, J.; Belanto, J.J.; Verdine, V.; Cox, D.B.T.; Kellner, M.J.; Regev, A.; et al. RNA targeting with CRISPR-Cas13. Nature 2017, 550, 280–284.
- Gilbert, L.A.; Horlbeck, M.A.; Adamson, B.; Villalta, J.E.; Chen, Y.; Whitehead, E.H.; Guimaraes, C.; Panning, B.; Ploegh, H.L.; Bassik, M.C.; et al. Genome-scale CRISPR-mediated control of gene repression and activation. Cell 2014, 159, 647–661.
- Hazafa, A.; Mumtaz, M.; Farooq, M.F.; Bilal, S.; Chaudhry, S.N.; Firdous, M.; Naeem, H.; Ullah, M.O.; Yameen, M.; Mukhtiar, M.S.; et al. CRISPR/Cas9: A powerful genome editing technique for the treatment of cancer cells with present challenges and future directions. Life Sci. 2020, 263, 118525.
- Hirakawa, M.P.; Krishnakumar, R.; Timlin, J.A.; Carney, J.P.; Butler, K.S. Gene editing and CRISPR in the clinic: Current and future perspectives. Biosci. Rep. 2020, 40, BSR20200127.
- Xu, D.; Cai, Y.; Tang, L.; Han, X.; Gao, F.; Cao, H.; Qi, F.; Kapranov, P. A CRISPR/Cas13-based approach demonstrates biological relevance of vlinc class of long non-coding RNAs in anticancer drug response. Sci. Rep. 2020, 10, 1794.
