G-quadruplexes, a family of (thermodynamically and kinetically stable) tetraplex helices, are non-canonical secondary structures derived from guanine (G)-rich sequences of nucleic acids. G-quadruplexes were found to occur in functionally-important regions of the human genome, including the telomere tandem sequences, several proto-oncogene promoters and other regulatory regions, ribosomal DNA (rDNA), as well as mRNA sequences encoding for proteins with roles in tumorigenesis, thus establishing a clear connection between G-quadruplexes and known hallmarks of cancer. Stabilization of G-quadruplexes belonging to the above categories, by means of small-molecule intervention, has been correlated with a range of anticancer effects, which has led to classifying G-quadruplexes as novel potential targets in anticancer research. The most common ways in which G-quadruplexes are now understood to serve in an anticancer capacity are presented herein.
- G-quadruplex
- telomerase inhibition
- proto-oncogene promoters
- ribosomal DNA
1. Introduction
G-quadruplexes, a family of tetraplex helices, are non-canonical secondary structures derived from guanine (G)-rich sequences of nucleic acids and exhibiting remarkable thermodynamic and kinetic stability [1]. While G-quadruplexes form readily in vitro from single nucleic acid strands, their assembly and stabilization in vivo, where they may exist in equilibrium with a different type of structure (e.g., double-stranded DNA), has been suggested to require the function of protein chaperons [2].
The following organization is characteristic of a G-quadruplex assembly: Guanines from the participating sequence(s), in sets of four, are oriented in square planar quartets, driven by a network of Hoogsteen hydrogen bonds (Figure 1A); G-quartet stability is further enhanced by coordination of (monovalent) cations to guanine carbonyls (Figure 1A); and G-quartets accumulate atop each other due to π-π stacking, while interconnected by the sugar-phosphodiester backbone (Figure 1B,C) [3,4,5].
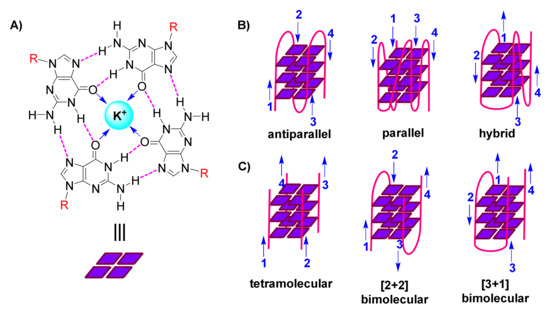
Figure 1. (A). Representation of a guanine(G)-quartet, highlighting the network of Hoogsteen hydrogen bonds (magenta), monovalent cation (cyan), dipole-cation interactions (blue arrows), and sites of connection to the sugar-phosphodiester backbone (R, red) (B). Cartoon representations of diverse unimolecular/intramolecular G-quadruplexes, with blue arrows indicating direction of each strand (numbered) (C). Cartoon representations of diverse intermolecular G-quadruplexes, with blue arrows indicating direction of each strand (numbered).
G-quadruplexes are polymorphic entities, as revealed by 3D structural studies, with their family comprising both unimolecular/intramolecular (Figure 1B) and intermolecular (Figure 1C) structures. These exhibit diversity in the lengths, sequences, folds and orientations of the loops that interconnect the participating strands, leading to classification of G-quadruplexes as parallel, antiparallel or hybrid (Figure 1B) [6,7,8].
Since the early days of this field, in attempting to answer the question whether G-quadruplexes are biologically relevant, algorithms have been devised and applied by various research teams, in order to predict possibility of occurrence of G-quadruplexes in the human and other genomes [9,10,11,12]. Genome-wide analyses have indicated a frequent occurrence of G-quadruplex-forming sequences in functional genomic regions, suggesting G-quadruplex association with telomere maintenance, replication, transcription and translation which, in turn, has led to suggestions of G-quadruplex-mediated regulatory mechanisms for these processes. The roles of G-quadruplexes in these processes are understood in much detail today [13].
Many of the >370,000 predicted G-quadruplex-forming sequences in humans [9,10] are traced in promoter regions of genes, close to transcription start sites [12]. Despite the fact that these predominantly exist in vivo in the form of double-stranded helices, their transient conversion to single-stranded is believed possible, in the course of replication, transcription and recombination. It can be achieved with the assistance of negative DNA supercoiling and conditions of molecular crowding, caused by protein binding, which favor folding into G-quadruplexes [14]. Moreover, the presence of tandem G-rich repeats in the human telomere [15,16], which is naturally single-stranded, energetically favors formation of multiple G-quadruplexes. On the other hand, RNAs containing G-quadruplex-forming motifs in their 5’-untranslated regions (5’-UTRs), estimated to be around 3000 in humans [17], are also single-stranded and readily fold into stable G-quadruplex structures.
Most G-quadruplex-related studies have been conducted ex vivo. However, accumulating experimental evidence is now providing proof of the in vivo occurrence of G-quadruplexes. An early study employing high-specificity antibodies against telomeric G-quadruplexes, raised by ribosome display, has achieved targeting of intermolecular antiparallel G-quadruplexes in the ciliate model organism Stylonychia [18]. More recent studies involving highly specific antibodies, have achieved visualization of G-quadruplexes in living human cancer cells [19,20] and tissues [21]. Also, over the last few years, there has been significant progress in the development of G-quadruplex-specific, small-molecule-based fluorescent probes and theranostics [22,23,24,25,26,27], which now find application as bioimaging agents to trace G-quadruplexes in a cellular context and expand our understanding on their functional roles in physiological processes, including those with consequences for cancer research.
The presence of G-quadruplex-forming motifs in key genomic DNA and RNA sequences, uniquely places them in position to regulate several cellular pathways. Importantly, many of these pathways are directly associated to well-established hallmarks of cancer [28]. Indicatively, G-quadruplexes have been correlated to chromosomal homeostasis, genome maintenance and integrity, apoptosis and survival, proto-oncogene and cancer protein expression and post-translational modifications [13]. G-quadruplex-forming sequences are often found amplified in certain cancers [29,30]. The realization of a strong link between G-quadruplexes and unprecedented anticancer mechanisms of action has leveraged G-quadruplex structures to therapeutic target status in oncology [31,32,33]. The physiological relevance and significance of G-quadruplexes in the context of cancer have been widely reviewed [34,35,36].
The putative roles of G-quadruplexes in prevention of cancer pathogenesis have been, for years, a major inspiration and drive for research efforts by many teams, with implications from a pharmacological perspective, for the design of small-molecule ligands targeting G-quadruplexes and aiming to induce G-quadruplex-mediated anticancer effects. A vast number of scaffolds have been proposed and new compounds designed and synthesized to address the task at hand, namely the binding (with high affinity and selectivity) and stabilization of G-quadruplexes in nucleic acid sequences of cancer relevance [36,37,38,39,40,41,42]. Cellular responses upon treatment of cells with G-quadruplex-targeting ligands have been correlated with the perceived function of these G-quadruplexes. In parallel, several methodologies for ascertaining the anticancer potential of G-quadruplex-stabilizing ligands have been described [43].
2. G-Quadruplex-Mediated Anticancer Mechanisms
2.1. Interference with Chromosomal Homeostasis & Telomerase-Mediated Telomere Elongation
The telomere is a region of repetitive nucleotide sequences at chromosomal ends which, via complexation with various nucleoproteins, folds into higher-order secondary structures, that play the role of a ‘cap’, protecting the chromosome from deterioration or fusion with other chromosomes [48]. The type of ‘cap’ secondary structure and the participating proteins exhibit variability between different species [2,49]. The existence of an intact ‘cap’ also prevents improper activation of DNA damage-response pathways [50].
G-quadruplexes occur in high concentrations in telomeres [19,51], due to the high guanine content of the telomere tandem sequence (TTAGGG in vertebrates) and are, in fact, capable of protecting genome integrity in cases where normal telomeric ‘caps’ are compromised [52]. In vitro studies have shown telomeric G-quadruplexes to interact with human proteins TRF2, EWS and FUS, which can co-bind the long non-coding RNA TERRA [53,54,55]. The simultaneous binding of telomeric and TERRA G-quadruplexes causes recruitment of histone methyltransferases by FUS, thus providing an association with telomere heterochromatin maintenance [54].
Stabilization of G-quadruplexes in the telomere during DNA replication could generate problems. Loss of telomeric G-quadruplex-interacting proteins, such as the CST cluster [56] and RTEL1 helicase [57], results in telomere shortening and fragility, and affords altered rates of replication [58]. The addition of G-quadruplex-stabilizing ligands was shown to exacerbate this situation [56].
Importantly, telomerase, a reverse transcriptase that is over-expressed in about 85% of cancer cells [59], stem cells and germline cells, is responsible for providing genomic stability by elongating the protruding 3’ single-stranded G-rich overhang at the ends of telomeres. For this extension to be permitted, base pairing needs to take place between the G-rich overhang and a RNA template carried by telomerase to encode the telomeric repeat sequence [60]. Elongation, which counteracts telomere shortening, may be inhibited by the formation of G-quadruplexes in telomeric sequences [61]. This is a result of hindered access of telomerase to the telomere sequence, caused by formation of antiparallel intramolecular G-quadruplexes (Figure 2A). However, alternative intermolecular parallel G-quadruplexes may also form, which can be partially resolved by telomerase in vitro, allowing the extension to proceed [62]. Evidence from Saccharomyces cerevisiae indicates a co-localization of parallel G-quadruplexes in the telomere with telomerase [62,63]. On the other hand, the POT1-TPP1 protein complex, responsible for recruitment of telomerase to the telomeres, is capable of destabilizing G-quadruplexes [64,65]. Recent evidence shows the importance of G-quadruplex formation in a POT1-TPP1 mediated DNA synthesis [66]. Finally, telomerase activity may be affected by the 5’ end unfolding of its RNA component, caused by a small molecule [67].
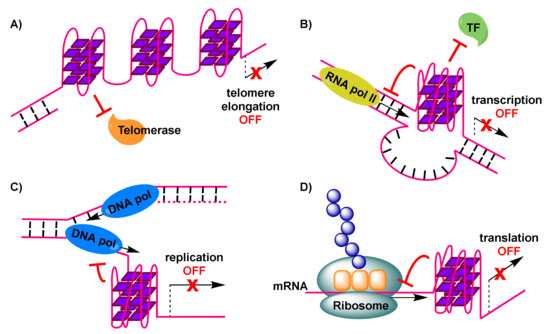
Figure 2. Formation of G-quadruplexes impacting physiological processes, with anticancer consequences: (A) G-quadruplexes in the telomere impose hindrance to telomerase, preventing elongation of the telomere and triggering DNA damage response signals. (B) G-quadruplex in oncogene-promoter region dislocates transcription factors and down-regulates RNA polymerase-mediated transcription of (onco)genes. (C) G-quadruplex in DNA undergoing replication stalls replication fork progression and leads to replicative stress, resulting in double strand breakpoints. (D) G-quadruplex in mRNA interferes with translation and formation of cancer proteins.
A vast number of ligands to stabilize telomeric G-quadruplexes in cancer cells have been described, despite the natural role of G-quadruplexes in telomerase-mediated telomere elongation not being fully elucidated. A resulting inhibition of telomerase activity upon addition of such ligands has been reported [68], while several ligands are able to displace members of the telomere protection complex shelterin, resulting in telomere damage and cell death [59,69]. While an alternative path of telomere elongation may be promoted upon G-quadruplex-imposed replication stress in certain cancer cells [70], presence of a G-quadruplex-stabilizing ligand may still result in cell death [71].
2.2. Transcriptional Regulation of Proto-Oncogene Promoters
The early detection, by means of applying computational predictive algorithms [12], of G-quadruplex-forming motifs in the promoter regions of several known proto-oncogenes [72], has indicated that G-quadruplexes are over-represented in these regions and may, in fact, possess regulatory roles with regard to the expression levels of oncogenes. Additional efforts have been successful in mapping G-quadruplex structures in chromatin to regulatory regions found adjacent to the transcription start sites of several of these genes in humans [30,73].
A number of in vitro studies applying small-molecule G-quadruplex-targeted ligands as agents inducing stabilization of G-quadruplexes in proto-oncogene promoter regions have demonstrated an ensuing reduction in oncogene transcription levels. Examples include transcriptional regulation of MYC, KRAS, KIT, BCL2 and VEGF [72,74,75,76,77]. However, explicit evidence of a link between G-quadruplexes and transcriptional control, coming from cellular studies, remains quite limited [78].
Indirect evidence of G-quadruplex impact on transcription of oncogenes is provided by the fact that certain transcription factors recognize G-quadruplex structures in vitro. Examples include recombinant nucleolin recognizing MYC [79], CNBP recognizing MYC [80] and SP1 recognizing ΚΙΤ [81]. This has led to the hypothesis that G-quadruplex-mediated mechanisms may be employed by nature for transcriptional regulation purposes.
To explain reduced expression levels of oncogenes, it has been suggested that G-quadruplex formation may impair initiation of transcription by preventing binding of RNA polymerase II and transcriptional machinery to the promoter transcription start site (Figure 2B) [74].
The formation of a G-quadruplex in the human telomerase reverse transcriptase gene (hTERT) has also been suggested to prevent binding of the gene repressor CCCTC binding factor, leading, in this case, to elevation of plasmid-encoded hTERT transcription [82].
2.3. Ribosomal DNA (rDNA) Transcription Inhibition
Ribosomal DNA (rDNA) is a GC-rich DNA sequence located in the nucleolus of cells, which encodes for ribosomal RNA. It contains more than 400 copies of the rRNA genes, organized in tandem arrays.
Ribosome biogenesis is under the control of multiple cellular signaling pathways, converging on the RNA polymerase I complex. RNA polymerase I is responsible for the transcription of rRNA genes and production of pre-rRNAs which, after maturation, will provide the main components for construction of the ribosome.
In cancer cells, proto-oncogene ‘gain-of-function’ and tumor-suppressor ‘loss-of-function’ mutations operate, leading to deregulated cellular signaling pathways, which in turn results in excessive ribosome biogenesis, required to support the rapid cell proliferation in tumors [83,84,85,86]. Given that the synthesis of rRNA by RNA polymerase I is considered the rate-limiting step in ribosome biogenesis [87], the interaction of rDNA with the RNA polymerase I protein complex could be a locus for anticancer intervention. Disruption of this interaction leads to arrest of ribosome biogenesis.
G-quadruplexes are believed to have a role in rDNA transcription. Specifically, G-quadruplexes may form transiently in the non-template strand in the course of rDNA transcription, and their occurrence prevents renaturation of the template DNA, assisting toward a dense arrangement of RNA polymerase I molecules on rRNA genes [88]. The formation of G-quadruplexes appears to be associated with their nanomolar-affinity interaction with nucleolin [89], an abundant nucleolar protein whose presence is essential for the progression of rDNA transcription [90]. Therefore, the disruption of G-quadruplex-nucleolin association, by means of interference with small-molecule ligands, is a way of inhibiting RNA polymerase I-mediated rDNA transcription, leading up to ribosome biogenesis suppression and eventually apoptosis of cancer cells.
2.4. Induction of Replication Stress Causing Genome Instability
Formation of G-quadruplexes in DNA sites during the transient opening of the double helix in the course of replication, has been implicated in increasing replication stress [91]. This is the result of obstruction caused to the progression of the replication forks (Figure 2C), leading to replication-fork collapse [92,93] and eventually the generation of double-strand breakpoints that cause genome instability and pose a threat to cell viability.
Via use of computational analyses of cancer databases, G-quadruplex formation has been associated with breakpoints in many cases in cancer cells, relevant to somatic copy-number alterations [94]. Stable G-quadruplexes were also found to be enriched in sites of somatic mutations, suggesting they may have roles as important determinants of mutagenesis [95]. G-quadruplex sequencing in the human genome has also revealed correlations of G-quadruplexes with gene amplifications, observed in cancer cells [29,30].
Evidence of genome instability due to G-quadruplex formation in the course of replication comes from elaborate studies in the model organisms Caenorhabditis elegans, Saccharomyces cerevisiae and Xenopus laevis, where the knock-out of a rescue system, namely helicases with the ability to resolve G-quadruplexes (such as DOG1, FANCJ and PIF1), renders the system prone to occurrence of DNA breakpoints [96,97,98,99,100,101]. These findings highlight the importance of helicases in cellular rescue mechanisms, as well as the relation between potential helicase ‘loss-of-function’ and genome instability.
2.5. Interference with Translation of Messenger RNA (mRNA) to Cancer Proteins
The bioinformatics discovery that G-quadruplex-forming motifs are prevalent in 5’-UTRs of RNAs [15], confirmed by spectroscopic studies on these sequences, has rendered such mRNA transcripts that encode for proteins with functional roles in cancer, attractive targets. The 5’-UTRs of mRNAs are located adjacent to translation initiation sites. Therefore, the formation of G-quadruplexes in 5’-UTRs of mRNA sequences (Figure 2D) may result in interference with mRNA translation [102] (e.g., potential formation of the ribosome at alternative, upstream start codons, thus preventing translation of the main open reading frame [103]), eventually depriving cancer cells of valuable proteins. An early prototype example, of interest to anticancer research, is the 5’-UTR of NRAS mRNA, where emergence of a G-quadruplex has been correlated with about 80% repression in protein levels in vitro, based on a luciferase reporter assay [17]. Many subsequent efforts, including studies in live cells, have identified additional G-quadruplex-forming sites in 5’-UTRs of the same and other mRNAs, which can be manipulated, by means of stabilization by appropriate small-molecule ligands, to achieve similar impact on translation.
G-rich sequences within mRNA coding regions are also encountered, however, at lower abundance compared to 5’-UTRs [104]. Upon G-quadruplex formation, they exhibit ability to stall translation, 6-7 nucleotides before the G-quadruplex [105].
The above findings, in addition to the identification of helicases capable of unwinding RNA G-quadruplexes [103], supports the notion that RNA G-quadruplexes may serve as a natural mechanism of regulating the expression levels of specific genes on a post-transcriptional level.
Small-molecule-based tools that offer the ability to modulate the stability of G-quadruplexes of this type, in a dose- and time-dependent manner, can be pharmacologically useful, especially given the single-stranded nature of mRNAs, which makes them more susceptible to modulation compared to dsDNAs.
This entry is adapted from the peer-reviewed paper 10.3390/molecules26040841
