C and N are the most important essential elements constituting organic compounds in plants. The shoots and roots depend on each other by exchanging C and N through the xylem and phloem transport systems. Complex mechanisms regulate C and N metabolism to optimize plant growth, agricultural crop production, and maintenance of the agroecosystem.
- C and N interactions
- fixation
- assimilation
- plant-microbiome interactions
- starch
- high-temperature
1. Metabolism and Transport of C and N
C and N are the most important essential elements in plants, animals, and microorganisms. They act as limiting factors for plant growth and crop yield, which makes their metabolism and transport important for agricultural practices. Plant roots absorb water and nutrients from the soil and transport them to the shoots via xylem vessels in the roots, stems, and leaves. Transpiration through the stomata in the leaves and root pressure are two driving forces the “pull” the water and dissolved nutrients upward from the roots to the leaves against the force of gravity. Plant roots generally absorb N in the form of ammonium and nitrate, the most dominant available N compounds in upland soil except for soil organic matter. Plant roots can absorb some organic N compounds, such as amino acids, although their contribution is generally low under natural conditions. Nitrate is the most abundant inorganic N form in upland fields because ammonium is readily oxidized to nitrate by nitrifying bacteria under aerobic conditions[1]Nitrate is absorbed through the nitrate transporters located in the plasma membranes of the cells and is reduced to nitrite by the enzyme nitrate reductase (NR) in the cytoplasm using NADH or NADPH as a reductant. The nitrite ions are incorporated into the plastids and further reduced to ammonium ions via enzyme nitrite reductase (NiR).
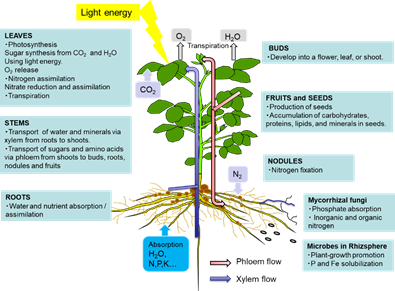
The leaves play a role in photosynthesis, which uses light energy to produce carbohydrates from the atmospheric CO2 and root-derived H2O while simultaneously releasing O2 as a by-product (Figure 1). Sucrose is the main form of photoassimilate transported from the mature leaves to the roots, symbionts, and growing sink organs, such as buds, flowers, and fruits, via the phloem. Leaves play an important role in N metabolic processes, such as nitrate reduction, assimilation, and amino acid transport, to support other organs.
Figure 1. Overview of the C and N flow among plant organs
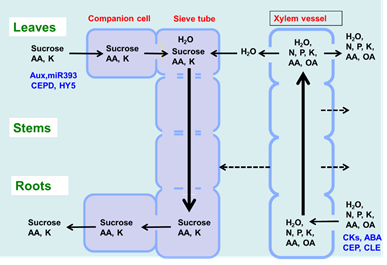
As shown in Figure 1, there are two routes by which materials are transported among plant organs in the vascular system of the xylem and phloem. The xylem vessels are a system of pipes made up of dead cells, through which water and absorbed nutrients, such as N, P, K, Ca, Mg, and minor elements, are transported from the roots to the shoots (Figure 2). The major driving forces for the upward movement against gravity are transpiration and root pressure. Photoassimilates (mainly sucrose), amino acids, and minerals such as K are transported bi-directionally from the leaves to the roots, buds, and fruits via a sieve tube in the phloem. This sieve tube is a channel of sieve-tube cells connected end-to-end, similar to xylem vessels. The gradient of the osmotic pressure between the source and sink organs is considered to be a driving force for the constant flow of phloem sap. However, the cooling of parts of the stems of some plant species causes a reversible decrease in phloem transport, suggesting that some biological processes, such as cytoplasmic streaming, may influence phloem transport.
Figure 2. Transport of C and N and regulatory signals through the phloem and xylem.
2. Regulation and Interaction of C and N Metabolism
The exchange of C and N between the shoots and roots through the xylem and phloem is crucial. N plays a significant role in C metabolism due to its function in protein synthesis. Likewise, C compounds are essential for N absorption, nitrate reduction, N2 fixation, and amino acid metabolism to generate C skeletons, metabolic energy, and reductants. Since a significant amount of fixed C is required to provide the C skeletons that act as acceptors for assimilating N into amino acids to form proteins and other nitrogenous compounds, a correlation between carbohydrate content increases and the downregulation of genes involved in photosynthesis and N metabolism has been reported[2]. N is a limiting factor for the growth and yield of most of crops[3]. When N availability is low, plant growth is stunted, and the leaves show chlorosis because of a decrease in the photosynthetic pigment chlorophyll. Besides under N deficiency conditions, the N in mature leaves is also mobilized to the growing parts and enhances the senescence of older leaves[4]. An increase in the N supply stimulates plant growth, making the plant taller, and also delays senescence. Furthermore, the N supply changes the plant morphology, as shown by an increase in the shoot to root dry weight ratio of both annual and perennial plant species [4]. However, excess N in the soil can be harmful to plants because it stimulates the overgrowth of vegetative organs and inhibits the growth of reproductive organs, ultimately decreasing crop yields[5][6]. The root architecture is modified by the levels and the placement of N in the soil. Yashima et al.[7] showed that soybean plants cultivated under an N-free nutrient solution have longer roots than plants with a nitrate supply.
The green revolution drastically increased cereal crop yields by up to two to four times by selecting semi-dwarf varieties, tolerant to the application of high amounts of chemical N fertilizers over original domestic varieties, ultimately showing overgrowth and lodging under high-N conditions. Increasing the productivity of the green revolution varieties of cereals increased environmental damage because of the increased application of N fertilizers and that it is vital to improve N-use efficiency (NUE) to solve environmental pollution problems[8]. The antagonistic activities and physical interactions of the rice growth-regulating factor4 transcription factor and the growth repressing DELLA (Aspartic acid-glutamic acid-leucine-leucine-alanine) protein are required for the homeostatic regulation of C and N metabolism and growth[8]. In wheat, bioactive gibberellins (GAs) promote plant growth by stimulating the degradation of DELLA proteins. In addition to playing a role in nutrition, C and N may act as signals to regulate nutrient absorption, assimilation, photosynthesis, and eventually plant growth and crop yield through the expression of related genes, enzymatic activities, and signal transduction networks[9]. Nitrate is absorbed by nitrate transporters (NRTs), and ammonium by ammonium transporters (AMTs), in the plasma membrane. The nitrate transporter, nitrate reductase, and glutamine synthetase genes were observed to be induced in sugar-depleted Arabidopsis supplied with extra exogenous sugar [10]. Three genes, nitrate transporters (LIN1/NRT2.1), glutamate receptor (GLR1.1), and a methyltransferase named oversensitive to sugar 1 (OSU1), were found to be involved in C–N signaling in Arabidopsis[11]. It was found that 2-phosphoglycolate (2-PG) derived from the oxygenase activity of Rubisco and 2-OG from the tricarboxylic acid (TCA) cycle act as C and N starvation signals, respectively.
Glutamate plays an essential role in the excitation of neurotransmitters in the mammalian nervous system via a family of ionotropic glutamate receptors (iGluRs) [12]. In 1998, it was found that plants contain a family of glutamate receptor-like (GLR) genes that are related to the mammalian iGluRs[13]. In animals, iGluRs operate as Glu- and Gly-gated non-selective cation channels allowing the uptake of K+, Na+, and Ca2+ into neurons, but plant GLR receptors have much broader amino acid specificity via Ala, Asn, Cys, Glu, Gly, and Ser[14]. However, despite significant recent progress in elucidating the functions and modes of action of the N sensory signaling systems, there is still much uncertainty about the extent to which they contribute to the process by which plants monitor their N status.
3. C and N metabolism through plant-microorganism interactions: mutualistic, pathogenic, and microbial volatile compounds
Plants release substantial amounts of complex photosynthetically derived C (20% to 50%) as exfoliates and root exudates (e.g., organic acids, flavonoids) into the rhizosphere [15], an input that plays a crucial factor in the increasing microbial abundance and activity in the rhizosphere compared to bulk soil. The extent to which this C flow (together with N assimilation and partitioning) is integrated into root and rhizosphere functions is of great interest for both basic (model plants and ecology) and applied sciences to increase crop yield and engage in plant disease control and/or bioremediation, thereby meeting our dramatically increasing demand for food. In this context, microorganisms have strong impacts on the organization and functioning of C and N metabolism in plants. According to their lifestyles (their involvement in the plant host), microorganisms can be divided into two groups, detrimental pathogen species (biotrophic to necrotrophic) or, more commonly, those with neutral or mutualistic interactions (e.g., below-ground, such as mycorrhiza, plant growth-promoting rhizobacterias (PGPRs), and rhizobia and above-ground, such as endophytes or epiphytes). In this context, the types of plant–-microbe interactions encompass competition, commensalism, mutualism, and parasitism, which can affect C and N metabolism. Both mutualistic and pathogenic microbes can colonize either sink or source organs and interfere with the source–sink balance due to their required sugar supply from host plants to the heterotrophic colonizing agent.
The beneficial plant–microbe interactions positively affecting plant growth, obtained via PGPR, pseudomonas, bacilli, trichoderma, diazotrophs, arbuscular mycorrhizal fungi (AMF), phosphate-solubilizing fungi, and bacteria or cellulose-degrading bacteria, are dependent on many external factors, including photosynthesis activity, plant size, and soil conditions [16][17]. C handling is a fundamental aspect of the mycorrhizal symbiosis. AMF derive most of their C from the host plant by increasing plant biomass and photosynthesis and directing the flow of a significant fraction of the host plant’s photoassimilates. Between 20% to 40% of the photoassimilates in host plants flow to mycorrhizal root systems (sink) to support these beneficial interactions[18]. In exchange, the AMF extraradical mycelium improves P acquisition by plants[19][20][21][22]; this P is used for energy supply, regeneration of the CO2 acceptor RuBP, and regulating the ratio of starch: Sucrose biosynthesis [23][24][25]. Moreover, mycorrhizal hyphae are thought to take up ammonium, nitrate, amino acids, and phosphate from the soil solution[26][27]. Besides, several studies on legume–rhizobia symbiosis, such as that in PGPR, demonstrated the importance of reduced C from plant photosynthesis in the form of carbohydrates, as well as amino acid and organic acid resources for the symbiotic N2-fixation that occurs in the root nodules[28][29]. The plant–microbe interactions may involve PGPR-triggered changes in plant sugar transport and C and N metabolism, which could lead to a regulated pool of sugars available for the PGPR and contribute to establishing and maintaining plant–PGPR symbiosis and its positive effects on plant fitness[30][31]. As N2-fixation enhances the leaf N mass fraction, it should also stimulate the plant photosynthesis rate by increasing RuBP activity and electron transport rates[32]. Microbes also synthesize and emit many volatile compounds (VCs) that can exert either inhibitory or stimulatory effects on plant growth by targeting the biochemical reactions that take place into the living cells[33][34][35][36]. depending on the microbial culture conditions, volatile emissions from some beneficial rhizosphere bacteria and fungi can promote plant growth[37][38][39][40][41]. Microbial VCs can promote changes in plants’ photosynthetic capacity, C metabolism, and transitions from source to sink status in photosynthetic tissues[42][43][44]. Moreover, microbial VCs have also been shown to increase root biomass and architecture, despite the aerial part [40][45][46].
In contrast to the situation in mutualistic symbiosis, biotrophic pathogens, including fungi, bacteria, and viruses, hijack host cells plants to suppress host immunity and take advantage of their nutrients, mainly sugars, for surviving and reproducing without returning any benefits to the host. The interactions between plants and biotrophic fungi (e.g., rust and powdery mildew) are cited more often as models for the study of pathogen-related modifications of C and N metabolism and partitioning. In the same line, plant–biotrophic virus interactions cause profound and dynamic modulations of the host’s primary metabolism. These perturbations include soluble sugar and starch accumulation at the infection sites, as well as reduced photosynthetic capacity indicative of a source to sink conversion[47]. Viruses exploit the assimilate transport system for their long-distance transport by interacting with the viral proteins to host factors and components of the long-distance transport machinery. In exchange, by activating the genes involved in carbohydrate catabolism cascades, such as glycolysis and the TCA cycle, plants use C and N metabolic pathways not only as a source of energy to drive extensive defense responses but also as a source of signaling molecules to trigger defense responses. In this line, as an inhibitory effect, VCs function as suppressors of immune responses, likely leading to the direct or indirect involvement of reactive oxygen species (ROS) as a signal leading to chlorosis. It has also been described that some VCs, via the primary transcriptional response, negatively affect the biological membranes represented by transport systems for sugar and amino acid permeability[48].
4. Carbon and Nitrogen Metabolism under Environmental Stresses
The most remarkable end-product of carbohydrates fixated by photosynthesis is the starch in plastids. Starch is a characteristic storage substance in plants that is not synthesized in other organisms. Starch is also the most important carbohydrate in the human diet. Starch is composed of amylose and amylopectin, which are glucose homopolymers and appear as semi-crystalline granules in plastids. Starch biosynthesis is performed with at least four enzymes: ADP-glucose pyrophosphorylase (AGPase), starch synthase (SS), starch-branching enzyme (BE), and starch-debranching enzyme (DBE). There are several proposed pathways for starch biosynthesis in plastids [49][50]. All starch synthesizing enzymes are encoded in the nuclear DNA. Nuclear-encoded plastidial proteins are generally synthesized in the cytosol and post-translationally imported into the organelle. The precursor proteins of starch synthesizing enzymes possess an N-terminal presequence called a transit peptide[51][52][53][54], which is necessary for, and also sufficient for, plastidial targeting and translocation initiation. The transit peptide is caught and interacts with the translocon at the outer envelope of the chloroplast (TOC) complex; then, protein import across the inner envelope is facilitated by the translocon at the inner envelope of chloroplast (TIC) complex[55][56][57]. After import, processing and folding of the precursor protein take place inside the plastid. Starch biosynthesis is regulated at both the transcriptional and posttranslational levels. On the contrary, starch degradation has been intensively investigated in germinating cereal seeds; α-amylase, debranching enzyme (R-enzyme), and α-glucosidase are involved in the breakdown process[58][59]. It has also been shown that α-amylase significantly contributes to the mechanism of starch breakdown in living cells of cereals[60][61][62]. It is worth noting that starch metabolism-related enzymes transport, localize, and functionalize plastids via diverse routes[63].
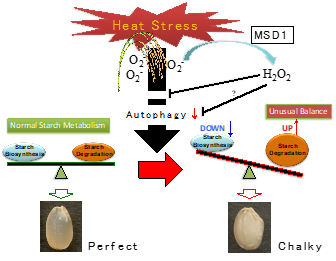
Diversification of the starch biosynthesis pathways occurring in both autotrophic and heterotrophic organs is likely finely regulated in response to the environmental inputs. Recently, extremely high temperatures during the ripening season of rice caused a decrease in yield and grain quality. It was reported that heat stress causes and stimulates grain chalkiness, which refers to damaged grain with loosely packed abnormal starch granules [64][65][66][67][68]. Severe grain chalkiness can lead to reductions in grain weight, resulting in yield losses. A proposed model of grain chalking under high-temperature stress is illustrated in Figure 3.
Figure 3. A proposed model of grain chalking under high-temperature stress.
This entry is adapted from the peer-reviewed paper 10.3390/ijms22010318
References
- Beevers, L. Nitrogen Metabolism in Plants; Edwward Arnold: London, UK, 1976.
- Kurusu, T.; Koyano, T.; Hanamata, S.; Kubo, T.; Noguchi, Y.; Yagi, C.; Natgata, N.; Yamanoto, T.; Ohnishi, T.; Okazaki, Y.; et al. OsATG7 is required for autophagy-dependent lipid metabolism in rice postmeiotic anther development. Autophagy 2014, 10, 878–888.
- Ohyama, T.; Minagawa, R.; Ishikawa, S.; Yamamoto, M.; Hung, N.V.P.; Ohtake, N.; Sueyoshi, K.; Sato, T.; Nagumo, Y.; Takahashi, Y. Soybean seed production and nitrogen nutrition. In A Comprehensive Survey of International Soybean Research-Genetics, Physiology, Agronomy and Nitrogen Relationships; Board, E., Ed.; InTech: Rijeka, Croatia, 2013; pp. 115–157.
- Marshiner, H. Function of mineral nutrients: Macronutrients. In Mineral Nutrition of Higher Plants, 2nd ed.; Academic Press: Amsterdam, The Netherland, 2002; pp. 229–312.
- Albornoz, F. Crop responses to nitrogen overfertilization: A review. Sci. Hort. 2016, 205, 79–83.
- Wang, D.; Xu, Z.; Zhao, J.; Wang, Y.; Yu, Z. Excessive nitrogen application decreases grain yield and increases nitrogen loss in a wheat-soil system. Acta Agric. Scand. Sect. B Soil Plant Sci. 2011, 61, 681–692.
- Yashima, H.; Fujikake, H.; Sato, T.; Ohtake, N.; Sueyoshi, K.; Ohyama, T. Systemic and local effects of long-term application of nitrate on nodule growth and N2 fixation in soybean (Glycine max [L.] Merr.). Soil Sci. Plant Nutr. 2003, 49, 825–834.
- Li, S.; Tian, Y.; Wu, K.; Ye, Y.; Yu, J.; Zhang, J.; Liu, Q.; Hu, M.; Li, H.; Tong, Y.; et al. Modulating plant growth–metabolism coordination for sustainable agriculture. Nature 2018, 560, 595–600.
- Goel, P.; Bhuria, M.; Kaushal, M.; Singh, A.K. Carbon: Nitrogen Interaction Regulates Expression of Genes Involved in N-Uptake and Assimilation in Brassica juncea L. PLoS ONE 2016, 11, e0163061.
- Gibon, Y.; Bläsing, O.E.; Palacios-Rojas, N.; Pankovic, D.; Hendriks, J.H.M.; Fisahn, J.; Möhne, M.; Günther, M.; Stitt, M. Adjustment of diurnal starch turnover to short days: Depletion of sugar during the night leads to a temporary inhibition of carbohydrate utilization, accumulation of sugars and post-translational activation of ADP-glucose pyrophosphorylase in the following light period. Plant J. 2004, 39, 847–862.
- Gent, L.; Forde, B.G. How do plants sense their nitrogen status? J. Exp. Bot. 2017, 68, 2531–2540.
- Forde, B.G. Glutamate signalling in roots. J. Exp. Bot. 2014, 65, 779–787.
- Lam, H.M.; Chiu, J.; Hsieh, M.H.; Meisel, L.; Oliveira, I.C.; Shin, M.; Coruzzi, G. Glutamate-receptor genes in plants. Nature 1998, 396, 125–126.
- Wang, R.; Okamoto, M.; Xing, X.; Crawford, N.M. Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1000 rapidly responding genes and new linkages to Glucose, Trehalose- 6-Phospate, Iron, and Sulfate Metabolism. Plant Physiol. 2003, 132, 556–567.
- Coleman, D.C.; Crossley, D.A., Jr.; Hendrix, P.F. Fundamentals of Soil Ecology, 2nd ed.; Elsevier: San Diego, CA, USA, 2004; ISBN 978-008-047-281-2.
- Mhlongo, M.I.; Piater, L.A.; Madala, N.E.; Labuschagne, N.; Dubery, I.A. The Chemistry of Plant–Microbe Interactions in the Rhizosphere and the Potential for Metabolomics to Reveal Signaling Related to Defense Priming and Induced Systemic Resistance. Front. Plant Sci. 2018, 9, 112.
- Adeniji, A.A.; Babalola, O.O.; Loots, D.T. Metabolomic applications for understanding complex tripartite plant-microbes interactions: Strategies and perspectives. Biotechnol. Rep. 2020, e00425.
- Bago, B.; Pfeffer, P.E.; Shachar-Hill, Y. Carbon metabolism and transport in arbuscular mycorrhizas. Plant Physiol. 2000, 124, 949–958.
- Baslam, M.; Garmendia, I.; Goicoechea, N. Arbuscular mycorrhizal fungi (AMF) improved growth and nutritional quality of greenhouse-grown lettuce. J. Agric. Food Chem. 2011, 59, 5504–5515.
- Baslam, M.; Garmendia, I.; Goicoechea, N. Elevated CO2 may impair the beneficial effect of arbuscular mycorrhizal fungi (AMF) on the mineral and phytochemical quality of lettuce. Ann. Appl. Biol. 2012, 161, 180–191.
- Baslam, M.; Pascual, I.; Sanchez-Diaz, M.; Goicoechea, N. Can Arbuscular Mycorrhizal Fungi (AMF) be Effective Tools for Improving the Nutritional Quality of Crops? In Findings from a Worldwide Consumed Vegetable: Lettuce. Beneficial Plant-Microbial Interactions: Ecology and Applications Science Publishers; CRC Press: Boca Raton, FL, USA, 2013; Volume 16, pp. 388–412. ISBN 978-042-907-374-8.
- Baslam, M.; Antolin, M.C.; Gogorcena, Y.; Muñoz, F.; Goicoechea, N. Changes in alfalfa forage quality and stem carbohydrates induced by arbuscular mycorrhizal fungi (AMF) and elevated atmospheric CO2. Ann. Appl. Biol. 2013, 164, 190–199.
- De Groot, C.C.; van den Boogaard, R.; Marcelis, L.F.M.; Harbinson, J.; Lambers, H. Contrasting effects of N and P deprivation on the regulation of photosynthesis in tomato plants in relation to feedback limitation. J. Exp. Bot. 2003, 54, 1957–1967.
- Rychter, A.M.; Rao, I.M. Role of phosphorus in photosynthetic carbon metabolism. In Handbook of Photosynthesis; Pessarakli, M., Ed.; Taylor and Francis Group, LLC: Tucson, AZ, USA, 2005; pp. 123–148. ISBN 978-131-537-213-6.
- Grimoldi, A.A.; Kavanova, M.; Lattanzi, F.A.; Schnyder, H. Phosphorus nutrition-mediated effects of arbuscular mycorrhiza on leaf morphology and carbon allocation in perennial ryegrass. New Phytol. 2005, 168, 435–444.
- Covindarajulu, M.; Pfeffer, P.E.; Jin, H.; Abubaker, J.; Douds, D.D.; Allen, J.W.; Bücking, H.; Lammers, P.J.; Shachar-Hill, Y. Nitrogen transfer in the arbuscular mycorrhizal symbiosis. Nature 2005, 435, 819–823.
- Bonfante, P.; Genre, A. Mechanisms underlying beneficial plant–fungus interactions in mycorrhizal symbiosis. Nat. Commun. 2010, 1, 48.
- Horst, I.; Welham, T.; Kelly, S.; Kaneko, T.; Sato, S.; Tabata, S.; Parniske, M.; Wang, T.L. TILLING mutants of Lotus japonicus reveal that nitrogen assimilation and fixation can occur in the absence of nodule-enhanced sucrose synthase. Plant Physiol. 2007, 144, 806–820. [Google Scholar] [CrossRef] [PubMed]
- Udvardi, M.; Poole, P.S. Transport and metabolism in legume-rhizobia symbioses. Ann. Rev. Plant Biol. 2013, 64, 781–805.
- Miotto-Vilanova, L.; Jacquard, C.; Courteaux, B.; Wortham, L.; Michel, J.; Clement, C.; Barka, E.A.; Sanchez, L. Burkholderia phytofirmans PsJN confers grapevine resistance against Botrytis cinerea via a direct antimicrobial effect combined with a better resource mobilization. Front. Plant Sci. 2016, 7, 1236.
- Hennion, N.; Durand, M.; Vriet, C.; Doidy, J.; Maurousset, L.; Lemoine, R.; Pourtau, N. Sugars en route to the roots. Transport, metabolism and storage within plant roots and towards microorganisms of the rhizosphere. Physiol. Plant 2019, 165, 44–57.
- Harley, P.C.; Thomas, R.B.; Reynolds, J.F.; Strain, B.R. Modelling photosynthesis of cotton grown in elevated CO2. Plant Cell Environ. 1992, 15, 271–282.
- Splivallo, R.; Novero, M.; Bertea, C.; Bossi, S.; Bonfante, P. Truffle volatiles inhibit growth and induce an oxidative burst in Arabidopsis thaliana. New Phytol. 2007, 175, 417–424.
- Tarkka, M.; Piechulla, B. Aromatic weapons: Truffles attack plants by the production of volatiles. New Phytol. 2007, 175, 381–383.
- Wenke, K.; Wanke, D.; Kilian, J.; Berendzen, K.; Harter, K.; Piechulla, B. Volatiles of two growth-inhibiting rhizobacteria commonly engage AtWRKY18 function. Plant J. 2012, 70, 445–459.
- Weise, T.; Kai, M.; Piechulla, B. Bacterial ammonia causes significant plant growth inhibition. PLoS ONE 2013, 8, e63538.
- Bailly, A.; Groenhagen, U.; Schulz, S.; Geisler, M.; Eberl, L.; Weisskopf, L. The inter-kingdom volatile signal indole promotes root development by interfering with auxin signaling. Plant J. 2014, 80, 758–771.
- Sánchez-López, Á.M.; Baslam, M.; De Diego, N.; Muñoz, F.J.; Bahaji, A.; Almagro, G.; Bahaji, A.; Almagro, G.; Ricarte-Bermejo, A.; García-Gómez, P.; et al. Volatile compounds emitted by diverse phytopathogenic microorganisms promote plant growth and flowering through cytokinin action: VCs from microbial phytopathogens promote growth. Plant Cell Environ. 2016, 39, 2592–2608.
- Sánchez-López, Á.M.; Bahaji, A.; de Diego, N.; Baslam, M.; Li, J.; Muñoz, F.J.; Almagro, G.; García-Gómez, P.; Ameztoy, K.; Ricarte-Bermejo, A.; et al. Arabidopsis responds to Alternaria alternata volatiles by triggering plastid phosphoglucose isomerase-independent mechanisms. Plant Physiol. 2016, 172, 1989–2001.
- García-Gómez, P.; Almagro, G.; Sánchez-López, Á.M.; Bahaji, A.; Ameztoy, K.; Ricarte-Bermejo, A.; Baslam, M.; Antolín, M.C.; Urdiain, A.; López-Belchi, M.D.; et al. Volatile compounds other than CO2 emitted by different microorganisms promote distinct posttranscriptionally regulated responses in plants. Plant Cell Environ. 2018, 42, 1729–1746.
- Ameztoy, K.; Baslam, M.; Sánchez-López, Á.M.; Muñoz, F.J.; Bahaji, A.; Almagro, G.; García-Gómez, P.; Almagro, G.; García-Gómez, P.; Humplík, J.F.; et al. Plant responses to fungal volatiles involve global posttranslational thiol redox proteome changes that affect photosynthesis. Plant Cell Environ. 2019, 9, 2627–2644.
- Zhang, H.; Xie, X.; Kim, M.S.; Kornyeyev, D.A.; Holaday, S.; Paré, P.W. Soil bacteria augment Arabidopsis photosynthesis by decreasing glucose sensing and abscisic acid levels in planta. Plant J. 2008, 56, 264–273.
- Ezquer, I.; Li, J.; Ovecka, M.; Baroja-Fernández, E.; Muñoz, F.J.; Montero, M.; Díaz de Cerio, J.; Hidalgo, M.; Sesma, M.T.; Bahaji, A.; et al. Microbial volatile emissions promote accumulation of exceptionally high levels of starch in leaves in mono- and di-cotyledonous plants. Plant Cell Environ. 2010, 51, 1674–1693.
- Li, J.; Ezquer, I.; Bahaji, A.; Montero, M.; Ovecka, M.; Baroja-Fernández, E.; Muñoz, F.J.; Merida, A.; Almagro, G.; Hidalgo, M.; et al. Microbial volatiles induced accumulation of exceptionally high levels of starch in Arabidopsis leaves is a process involving NTRC and starch synthases class III and IV. Mol. Plant-Microbe 2011, 24, 1165–1178.
- Piechulla, B.; Lemfack, M.C.; Kai, M. Effects of discrete bioactive. microbial volatiles on plants and fungi. Plant Cell Environ. 2017, 40, 2042–2067.
- Schenkel, D.; Maciá-Vicente, J.G.; Bissell, A.; Splivallo, R. Fungi Indirectly Affect Plant Root Architecture by Modulating Soil Volatile Organic Compounds. Front. Microbiol. 2018, 9, 1847.
- Beimalt, S.; Sonnwald, U. Plant-microbe interactions to probe regulation of plant carbon metabolism. J. Plant Physiol. 2006, 163, 307–318.
- Etschmann, M.M.W.; Bluemke, W.; Sell, D.; Schrader, J. Biotechnological production of 2-phenylethanol. Appl. Microbiol. Biotechnol. 2002, 59, 1–8.
- Baroja-Fernández, E.; Muñoz, F.J.; Zandueta-Criado, A.; Morán-Zorzano, M.T.; Viale, A.M.; Alonso-Casajús, N.; Pozueta-Romero, J. Most of ADP·glucose linked to starch biosynthesis occurs outside the chloroplast in source leaves. Proc. Natl Acad. Sci. USA 2004, 101, 13080–13085.
- Bahaji, A.; Baroja-Fernández, E.; Sánchez-López, Á.M.; Muñoz, F.J.; Li, J.; Almagro, G.; Montero, M.; Pujol, P.; Galarza, R.; Kaneko, K.; et al. HPLC-MS/MS analyses show that the near-starchless aps1 and pgm leaves accumulate wild type levels of ADPglucose: Further evidence for the occurrence of important ADPglucose biosynthetic pathway(s) alternative to the pPGI-pPGM-AGP pathway. PLoS ONE 2014, 9, e104997.
- Bruce, B.D. Chloroplast transit peptides: Structure, function and evolution. Trends. Cell Biol. 2000, 10, 440–447.
- Zhang, X.P.; Glaser, E. Interaction of plant mitochondrial and chloroplast signal peptides with the Hsp70 molecular chaperone. Trends. Plant Sci. 2002, 7, 14–21.
- Lee, D.W.; Kim, J.K.; Lee, S.; Choi, S.; Kim, S.; Hwang, I. Arabidopsis nuclear-encoded plastid transit peptides contain multiple sequence subgroups with distinctive chloroplast-targeting sequence motifs. Plant Cell 2008, 20, 1603–1622.
- Li, H.-m.; Teng, Y.-S. Transit peptide design and plastid import regulation. Trends Plant Sci. 2013, 18, 360–366.
- Inoue, H.; Li, M.; Schnell, D.J. An essential role for chloroplast heat shock protein 90 (Hsp90C) in protein import into chloroplasts. Proc. Natl. Acad. Sci. USA 2013, 110, 3173–3178.
- Kikuchi, S.; Bédard, J.; Hirano, M.; Hirabayashi, Y.; Oishi, M.; Imai, M.; Takase, M.; Ide, T.; Nakai, M. Uncovering the protein translocon at the chloroplast inner envelope membrane. Science 2013, 339, 571–574.
- Paila, Y.D.; Richardson, L.G.; Schnell, D.J. New insights into the mechanism of chloroplast protein import and its integration with protein quality control, organelle biogenesis and development. J. Mol. Biol. 2015, 427, 1038–1060.
- Akazawa, T.; Mitsui, T.; Hayashi, M. Recent progress in α-amylase biosynthesis. In The Biochemistry of Plants; Preiss, J., Ed.; Academic Press: New York, NY, USA, 1988; Volume 14, pp. 465–492.
- Beck, E.; Ziegler, P. Biosynthesis and degradation of starch in higher plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1989, 40, 95–117.
- Perez, C.M.; Palmiano, E.P.; Baun, L.C.; Juliano, B.O. Starch metabolism in the leaf sheaths and culm of rice. Plant Physiol. 1971, 47, 404–408.
- Asatsuma, S.; Sawada, C.; Itoh, K.; Okito, M.; Kitajima, A.; Mitsui, T. Involvement of α-amylase I-1 in starch degradation in rice chloroplasts. Plant Cell Physiol. 2005, 46, 858–869.
- Asatsuma, S.; Sawada, C.; Kitajima, A.; Asakura, T.; Mitsui, T. α-Amylase affects starch accumulation in rice grain. J. Appl. Glycosci. 2006, 53, 187–192.
- Jarvis, P. Targeting of nucleus-encoded proteins to chloroplasts in plants. New Phytol. 2008, 179, 257–285.
- Tashiro, T.; Wardlaw, I.F. The effect of high temperature on the accumulation of dry matter, carbon and nitrogen in the kernel of rice. Aust. J. Plant Physiol. 1991, 18, 259–265.
- Lisle, A.J.; Martin, M.; Fitzgerald, M.A. Chalky and translucent rice grains differ in starch composition and structure and cooking properties. Cereal Chem. 2000, 77, 627–632.
- Kim, S.S.; Lee, S.E.; Kim, O.W.; Kim, D.C. Physicochemical characteristics of chalky kernels and their effects on sensory quality of cooked rice. Cereal Chem. 2000, 77, 376–379.
- Singh, N.; Sodhi, N.S.; Kaur, M.; Saxena, S.K. Physicochemical, morphological, thermal, cooking and textural properties of chalky and translucent rice kernels. Food Chem. 2003, 82, 433–439.
- Ishimaru, T.; Horigane, A.K.; Ida, M.; Iwasawa, N.; San-oh, Y.A.; Nakazono, M.; Nishizawa, N.K.; Masumura, T.; Kondo, M.; Yoshida, M. Formation of grain chalkiness and changes in water distribution in developing rice caryopses grown under hightemperature stress. J. Cereal Sci. 2009, 50, 166–174.