The Actinobacteria phylum represents one of the most numerous and heterogeneous groups of microorganisms present in nature.
- Bifidobacteria
1. History and Taxonomy
The Actinobacteria phylum represents one of the most numerous and heterogeneous groups of microorganisms present in nature[1][2]. These Gram-positive bacteria are characterized by a high GC genome content ranging from 51% to more than 70%, and exhibit different morphologies, including unicellular rods or Y-shaped rods, and complex multicellular consortia[1][2]. Furthermore, these bacteria are able to produce bioactive natural compounds and these features are reflected in their ability to adapt to several quite distinct ecosystems such as various terrestrial environments, as well as the bodies of mammals and birds[1][2]. In fact, this phylum includes pathogens (e.g., Mycobacterium spp., Nocardia spp., Tropheryma spp., Corynebacterium spp., and Propionibacterium spp.), soil inhabitants such as Streptomyces spp., plant commensals (e.g., Leifsonia spp.), nitrogen-fixing symbionts (Frankia), and human gut inhabitants (Bifidobacterium spp.)[1][2].
The genus Bifidobacterium belongs to the Bifidobacteriaceae family, Bifidobacteriales order, and these bacteria were isolated, for the first time, from feces of a breast-fed infant by Tissier in 1899[3]. They represent nonmotile, anaerobic, nonsporulating, saccharolytic bacteria with a bifid or multiple-branching rod morphology. Currently, the genus Bifidobacterium comprises 94 taxa, representing 82 species and 12 subspecies[4][5][6][7][8][9][10][11][12] (Table 1). In recent years, the phylogeny of the Bifidobacterium genus has been explored using different methods based on the sequencing of the 16S rRNA gene, by means of a multilocus approach, or the sequencing of several housekeeping genes (i.e., clpC, dnaJ, rpoC, xpf, dnaB, and purF)[13][14]. A comparative genomics analysis based on all 88 sequenced bifidobacterial type strains revealed the presence of 191 Bifidobacterium-specific clusters of orthologous genes (COGs) shared by these genomes, called the bifidobacterial core-genome[15]. Notably, the phylogenetic tree constructed by amino acid concatenation of these 191 bifidobacterial core-genome genes revealed the existence of 10 different phylogenetic groups, encompassing Bifidobacterium adolescentis, Bifidobacterium boum, Bifidobacterium pullorum, Bifidobacterium asteroides, Bifidobacterium longum, Bifidobacterium psychraerophilum, Bifidobacterium bifidum, Bifidobacterium pseudolongum, Bifidobacterium bombi, and Bifidobacterium tissieri groups[15]. These groups partially correlate with the ecological niches from which the representative species were isolated. For example, members of the B. tissieri group are common inhabitants of the microbiota of tamarin and those of the B. pullorum group are characteristic of birds. According to this, members of the B. adolescentis group (Bifidobacterium catenulatum, Bifidobacterium pseudocatenulatum, and B. adolescentis strains), the B. longum group (Bifidobacterium breve and B. longum strains), the B. pseudolongum group (especially Bifidobacterium animalis subsp. lactis strains), and the B. bifidum group (B. bifidum strains) are typical colonizers of the human intestinal tract or are commercially exploited as probiotic strains (Figure 1).
Table 1. Bifidobacterium (sub)species recognized as reference strains (type strains).
|
Bifidobacterium Strains |
Isolation |
References |
|
B. actinocoloniiforme DSM 22766 |
Bumblebee digestive tract |
[16] |
|
B. adolescentis ATCC 15703 |
Intestine of human adult |
[17] |
|
B. aemilianum XV10 |
Carpenter bee digestive tract |
[5] |
|
B. aerophilum DSM 100689 |
Feces of cotton-top tamarin |
[18] |
|
B. aesuclapii DSM 26737 |
Feces of baby common marmoset |
[19] |
|
B. angulatum LMG 11039 |
Feces of human |
[20] |
|
B. animalis subsp. animalis LMG 10508 |
Feces of rat |
[21] |
|
B. animalis subsp. lactis DSM 10140 |
Fermented milk |
[22] |
|
B. anseris LMG 30189 |
Feces of domestic goose |
[7] |
|
B. apri DSM 100238 |
Digestive tract of wild pig |
[23] |
|
B. aquikefiri LMG 28769 |
Water kefir |
[24] |
|
B. asteroides LMG 10735 |
Hindgut of honeybee |
[25] |
|
B. avesanii DSM 100685 |
Feces of cotton-top tamarin |
[18] |
|
B. biavatii DSM 23969 |
Feces of tamarin |
[26] |
|
B. bifidum LMG 11041 |
Feces of breast-fed infant |
[3] |
|
B. bohemicum DSM22767 |
Bumblebee digestive tract |
[16] |
|
B. bombi DSM 19703 |
Bumblebee digestive tract |
[27] |
|
B. boum LMG 10736 |
Rumen of bovine |
[28] |
|
B. breve LMG 13208 |
Infant stool |
[17] |
|
B. callimiconis LMG 30938 |
Feces of Goeldi’s marmoset |
[6] |
|
B. callitrichidarum DSM 103152 |
Feces of emperor tamarin |
[29] |
|
B. callitrichos DSM 23973 |
Feces of common marmoset |
[26] |
|
B. canis DSM105923 |
Feces of dog |
[10] |
|
B. castoris LMG 30937 |
Feces of beaver |
[6] |
|
B. catenulatum LMG 11043 |
Adult intestine |
[30] |
|
B. catenulatum subsp. kashiwanohense DSM21854 |
Infant feces |
[31] |
|
B. catulorum DSM103154 |
Feces of common marmoset |
[32] |
|
B. cebidarum LMG31469 |
Feces of golden-headed tamarin |
[9] |
|
B. choerinum LMG 10510 |
Feces of piglet |
[28] |
|
B. commune LMG28292 |
Bumblebee gut |
[33] |
|
B. coryneforme LMG 18911 |
Hindgut of honeybee |
[25] |
|
B. criceti LMG 30188 |
Feces of European hamster |
[7] |
|
B. crudilactis LMG 23609 |
Raw cow milk |
[34] |
|
B. cuniculi LMG 10738 |
Feces of rabbit |
[28] |
|
B. dentium LMG 11045 |
Oral cavity |
[30] |
|
B. dolichotidis LMG 30941 |
Feces of Patagonian mara |
[6] |
|
B. eulemuris DSM 100216 |
Feces of black lemur |
[35] |
|
B. felsineum DSM103139 |
Feces of cotton-top tamarin |
[11] |
|
B. gallicum LMG 11596 |
Adult intestine |
[36] |
|
B. goeldii LMG 30939 |
Feces of Goeldi’s marmoset |
[6] |
|
B. hapali DSM 100202 |
Feces of baby common marmoset |
[37] |
|
B. imperatoris LMG 30297 |
Feces of emperor tamarin |
[7] |
|
B. indicum LMG 11587 |
Insect |
[25] |
|
B. italicum LMG 30187 |
Feces of European rabbit |
[7] |
|
B. jacchi DSM 103362 |
Feces of baby common marmoset |
[38] |
|
B. lemurum DSM 28807 |
Feces of ring-tailed lemur |
[39] |
|
B. leontopitechi LMG 31471 |
Feces of Goeldi’s monkey |
[9] |
|
B. longum subsp. infantis ATCC 15697 |
Intestine of infant |
[17] |
|
B. longum subsp. longum LMG 13197 |
Adult intestine |
[17] |
|
B. longum subsp. suis LMG 21814 |
Feces of pig |
[40] |
|
B. magnum LMG 11591 |
Feces of rabbit |
[30] |
|
B. margollesii LMG 30296 |
Feces of pygmy marmoset |
[7] |
|
B. meryciucm LMG 11341 |
Rumen of bovine |
[41] |
|
B. minimum LMG 11592 |
Sewage |
[42] |
|
B. mongoliense DSM 21395 |
Fermented mare’s milk |
[43] |
|
B. moukabalense DSM 27321 |
Feces of gorilla |
[44] |
|
B. myosotis DSM 100196 |
Feces of common marmoset |
[37] |
|
B. parmae LMG 30295 |
Feces of pygmy marmoset |
[7] |
|
B. platyrrhinorum SMA15 |
Feces of squirrel monkey |
[45] |
|
B. primatium DSM 100687 |
Feces of cotton-top tamarin |
[11] |
|
B. pseudocatenulatum LMG 10505 |
Infant feces |
[28] |
|
B. pseudolongum subsp. globosum LMG 11596 |
Rumen of bovine |
[46] |
|
B. pseudolongum subsp. pseudolongum LMG 11571 |
Feces of swine |
[21] |
|
B. psychraerophilum LMG 21775 |
Caecum of pig |
[47] |
|
B. pullorum subsp. gallinarum LMG 11586 |
Caecum of chicken |
[48] |
|
B. pullorum subsp. pullorum LMG 21816 |
Feces of chicken |
[8] |
|
B. ramosum DSM 100688 |
Feces of cotton-top tamarin |
[18] |
|
B. reuteri DSM 23975 |
Feces of common marmoset |
[26] |
|
B. rousetti BCRC 81136 |
Feces of Egyptian fruit bat |
[49] |
|
B. ruminantium LMG 21811 |
Rumen of bovine |
[41] |
|
B. pullorum subsp. saeculare LMG 14934 |
Feces of rabbit |
[50] |
|
B. saguini LMG 23967 |
Feces of tamarin |
[26] |
|
B. saimiriisciurei SMA1 |
Feces of squirrel monkey |
[45] |
|
B. saimirii LMG 30940 |
Feces of Bolivian saimiri |
[6] |
|
B. scaligerum DSM 103140 |
Feces of cotton-top tamarin |
[11] |
|
B. scardovii LMG 21589 |
Blood |
[51] |
|
B. simiarum DSM 103153 |
Feces of emperor tamarin |
[11] |
|
B. stellenboschense DSM 23968 |
Feces of tamarin |
[26] |
|
B. subtile LMG 11597 |
Sewage |
[42] |
|
B. porcinum LMG 21689 |
Feces of piglet |
[52] |
|
B. thermacidophilum LMG 21395 |
Anaerobic digester |
[53] |
|
B. termophilum JCM 7027 |
Rumen of bovine |
[21] |
|
B. tibiigranuli LMG 31086 |
Water kefir |
[54] |
|
B. tissieri DSM 100201 |
Feces of baby common marmoset |
[37] |
|
B. tsurumiense JCM 13495 |
Hamster dental plaque |
[55] |
|
B. vansinderenii LMG 30126 |
Feces of emperor tamarin |
[56] |
|
B. vespertilionis DSM 106025 |
Feces of Egyptian fruit bat |
[49] |
|
B. xylocopae DSM104955 |
Carpenter bee digestive tract |
[5] |
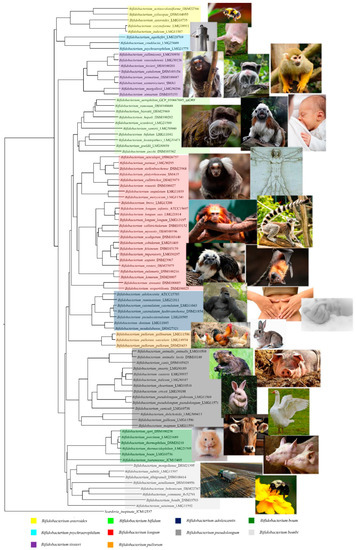
Figure 1. Phylogenetic tree of the Bifidobacterium genus based on the concatenation of 191 core amino acid sequence genes. The core genes-based tree shows the subdivision of the 10 phylogenetic groups of the Bifidobacterium genus represented with different colors. The phylogenetic tree was built by the neighbor-joining method with corresponding sequences of Scardovia inopinata JCM 12,537 being employed as outgroup. Bootstrap percentages above 50 are shown at node points, based on 100 replicates of the phylogenetic tree. The ecological origins of the various phylogenetic groups are highlighted beside the phylogenetic tree.
2. Ecology
Bifidobacteria also naturally occur in the gastrointestinal tract (GIT) of animals, such as nonhuman mammals, insects, and birds[5][6][7][8][9][10][11], while they have also been isolated from human blood[51], sewage[42], the oral cavity[55], and fermented milk[15]. In this context, it has been demonstrated that the ability of bifidobacteria to adapt to specific environments is species-dependent[4]. Until recently, scientific studies revealed that B. longum, B. adolescentis, B. pseudolongum, and B. bifidum species possess a cosmopolitan lifestyle[4], whereas other bifidobacterial species appear to be adapted to the GIT of particular animals (e.g., Bifidobacterium cuniculi for rabbits, Bifidobacterium angulatum for cows, and Bifidobacterium gallinarum for chickens) or the human gut (e.g., B. breve and B. longum species)[4][12]. However, recent ecological studies, based on Internally Transcribed Spacer (ITS) profiling, have revealed that the distribution of Bifidobacterium species is not host-specific[57][58]. For example, the B. breve species, which until that point had only been associated with the human gut, was shown to be present also in domesticated animals[57]. Furthermore, particular species, such as Bifidobacterium actinocoloniiforme, B. asteroides, Bifidobacterium bohemicum, B. bombi, and Bifidobacterium indicum, which were previously thought to be highly specialized to colonize the insect gut, were shown to be widely distributed among various mammalian hosts[58]. Notably, the distribution of bifidobacterial species in different ecological niches reinforces the idea that anthropogenic influences may have promoted such apparent horizontal transmission events.
The Bifidobacterium genus is one of the most abundant bacterial genera present in the human gut during the early stages of life[59][60][61] and these microorganisms are reported to be among the first bacterial colonizers of the newborn’s GIT[62]. It has been demonstrated that bifidobacteria may engage in vertical transmission that occurs between a mother and her newborn during birth and possibly through subsequent breastfeeding[63][64]. This fascinating phenomenon not only occurs in human beings[65] but also in other mammalian species[58]. In this context, some studies have shown how taxonomic classification of bifidobacteria present in the mother’s microbiota strongly correlates with that of the infant[66][67]. In particular, a study based on ITS-profiling and shotgun-metagenomics approaches has led to the identification of the species shared between a mother and her child[68]. In this study, the microbiota of a mother’s fecal and milk samples were assayed together with corresponding infant fecal samples collected at different time points. These analyses demonstrated that in some cases, identical bifidobacterial strains are shared in both mother’s and baby’s gut microbiota[68]. A B. breve strain and B. longum subsp. longum isolate were seen to be among the protagonists of vertical transmission from mother to child, being found both in the newborn’s meconium and in the fecal samples of the child for up to 90 days[68]. Several species of this genus are believed to have undergone specific genetic and metabolic adaptations in order to facilitate colonization of the infant gut, for example, the ability of certain bifidobacterial species and strains to metabolize specific oligosaccharides present in human milk[69]. Specifically, bifidobacterial species that are prevalent in the gut of infants include B. breve, B. longum subsp. infantis, B. longum subsp. longum, B. pseudocatenulatum, and B. bifidum[62], whereas B. adolescentis, B. catenulatum, B. pseudocatenulatum, and B. longum subsp. longum[70][71] are commonly occurring species in the adult intestine. In this context, it is not fully correct to consider the use of fecal material as a representation of the entire intestinal microbiota. In fact, the fecal microbiota consists not only of mucosal adherent members of the human GIT microbiota but also of transient bacteria derived from the diet or other environmental microbial contaminations[72]. Specifically, only a small number of bifidobacterial species (i.e., B. longum, B. adolescentis, B. breve, B. pseudocatenulatum, and B. pseudolongum) seem to be dominant in the examined biopsies, whereas certain other bifidobacterial species are restricted to a specific ecological niche (e.g., B. bifidum and B. pseudolongum)[73]. Analyses not only of human intestinal mucosal but also of fecal samples have shown that bifidobacterial distribution changes within ages, with a remarkable conservation in terms of species and strains in adults and children[73]. Furthermore, little is known about the diversity of bifidobacterial populations occurring between individuals and between different compartments of the GIT within the same individual[73].
The presence of bifidobacteria in the GIT has been associated with various health benefits, including the development of the immune system, protection against pathogens mediated through the process of competitive exclusion, and/or the production of metabolites such as short-chain fatty acids (SCFA) and vitamins[59][62][70][74]. Indeed, human-residential bifidobacteria (HRB) are also capable of producing folate, also known as vitamin B9 or B11, which is required for an efficient DNA replication, DNA repair/methylation, and synthesis of nucleotides, vitamins, and certain amino acids[75][76]. For these reasons, several bifidobacterial strains/species are used as active ingredients in a variety of so-called functional foods due to their perceived health-promoting or probiotic properties[2]. In this context, probiotic bifidobacterial strains belonging to B. longum and Bifidobacterium animalis subsp. lactis species are usually added to yogurt, other fermented milks, and, more recently, to cheese, which are the most popular probiotic foodstuffs at the moment[77][78]. Moreover, clinical studies have demonstrated that B. animalis ssp. lactis Bb-12, administered as probiotic adjunctive therapy, have beneficial effects in the case of infectious diarrhea caused by viruses or bacteria[79][80][81], decreasing the frequency or shortening the duration of the infection and increasing immune responses[81].
This entry is adapted from the peer-reviewed paper 10.3390/microorganisms9010008
References
- Van Bergeijk, D.A.; Terlouw, B.R.; Medema, M.H.; van Wezel, G.P. Ecology and genomics of Actinobacteria: New concepts for natural product discovery. Nat. Rev. Microbiol. 2020, 18, 546–558.
- Ventura, M.; Canchaya, C.; Tauch, A.; Chandra, G.; Fitzgerald, G.F.; Chater, K.F.; van Sinderen, D. Genomics of Actinobacteria: Tracing the evolutionary history of an ancient phylum. Microbiol. Mol Biol. Rev. 2007, 71, 495–548.
- Tissier, H. Recherches Sur la Flore Intestinale des Nourrissons: (état Normal et Pathologique). Ph.D. Thesis, G. Carré et C. Naud, Paris, France, 1900; p. 253.
- Alessandri, G.; Ossiprandi, M.C.; MacSharry, J.; van Sinderen, D.; Ventura, M. Bifidobacterial Dialogue With Its Human Host and Consequent Modulation of the Immune System. Front. Immunol. 2019, 10, 2348.
- Alberoni, D.; Gaggia, F.; Baffoni, L.; Modesto, M.M.; Biavati, B.; Di Gioia, D. Bifidobacterium xylocopae sp. nov. and Bifidobacterium aemilianum sp. nov., from the carpenter bee (Xylocopa violacea) digestive tract. Syst. Appl. Microbiol. 2019, 42, 205–216.
- Duranti, S.; Lugli, G.A.; Napoli, S.; Anzalone, R.; Milani, C.; Mancabelli, L.; Alessandri, G.; Turroni, F.; Ossiprandi, M.C.; van Sinderen, D.; et al. Characterization of the phylogenetic diversity of five novel species belonging to the genus Bifidobacterium: Bifidobacterium castoris sp. nov., Bifidobacterium callimiconis sp. nov., Bifidobacterium goeldii sp. nov., Bifidobacterium samirii sp. nov. and Bifidobacterium dolichotidis sp. nov. Int. J. Syst. Evol. Microbiol. 2019, 69, 1288–1298.
- Lugli, G.A.; Mangifesta, M.; Duranti, S.; Anzalone, R.; Milani, C.; Mancabelli, L.; Alessandri, G.; Turroni, F.; Ossiprandi, M.C.; van Sinderen, D.; et al. Phylogenetic classification of six novel species belonging to the genus Bifidobacterium comprising Bifidobacterium anseris sp. nov., Bifidobacterium criceti sp. nov., Bifidobacterium imperatoris sp. nov., Bifidobacterium italicum sp. nov., Bifidobacterium margollesii sp. nov. and Bifidobacterium parmae sp. nov. Syst. Appl. Microbiol. 2018, 41, 173–183.
- Trovatelli, L.D.; Crociani, F.; Pedinotti, M.; Scardovi, V. Bifidobacterium pullorum sp. nov.: A new species isolated from chicken feces and a related group of bifidobacteria isolated from rabbit feces. Arch. Microbiol. 1974, 98, 187–198.
- Duranti, S.; Lugli, G.A.; Viappiani, A.; Mancabelli, L.; Alessandri, G.; Anzalone, R.; Longhi, G.; Milani, C.; Ossiprandi, M.C.; Turroni, F.; et al. Characterization of the phylogenetic diversity of two novel species belonging to the genus Bifidobacterium: Bifidobacterium cebidarum sp. nov. and Bifidobacterium leontopitheci sp. nov. Int. J. Syst. Evol. Microbiol. 2020, 70, 2288–2297.
- Neuzil-Bunesova, V.; Lugli, G.A.; Modrackova, N.; Makovska, M.; Mrazek, J.; Mekadim, C.; Musilova, S.; Svobodova, I.; Spanek, R.; Ventura, M.; et al. Bifidobacterium canis sp. nov., a novel member of the Bifidobacterium pseudolongum phylogenetic group isolated from faeces of a dog (Canis lupus f. familiaris). Int. J. Syst. Evol. Microbiol. 2020, 70, 5040–5047.
- Modesto, M.; Puglisi, E.; Bonetti, A.; Michelini, S.; Spiezio, C.; Sandri, C.; Sgorbati, B.; Morelli, L.; Mattarelli, P. Bifidobacterium primatium sp. nov., Bifidobacterium scaligerum sp. nov., Bifidobacterium felsineum sp. nov. and Bifidobacterium simiarum sp. nov.: Four novel taxa isolated from the faeces of the cotton top tamarin (Saguinus oedipus) and the emperor tamarin (Saguinus imperator). Syst. Appl. Microbiol. 2018, 41, 593–603.
- Turroni, F.; van Sinderen, D.; Ventura, M. Genomics and ecological overview of the genus Bifidobacterium. Int. J. Food Microbiol. 2011, 149, 37–44.
- Philippe, H.; Douady, C.J. Horizontal gene transfer and phylogenetics. Curr. Opin. Microbiol. 2003, 6, 498–505.
- Ventura, M.; Canchaya, C.; Casale, A.D.; Dellaglio, F.; Neviani, E.; Fitzgerald, G.F.; van Sinderen, D. Analysis of bifidobacterial evolution using a multilocus approach. Int. J. Syst. Evol. Microbiol. 2006, 56, 2783–2792.
- Gabriele Andrea Lugli; Christian Milani; Sabrina Duranti; Giulia Alessandri; Francesca Turroni; Leonardo Mancabelli; Danilo Tatoni; Maria Cristina Ossiprandi; Douwe Van Sinderen; Marco Ventura; et al. Isolation of novel gut bifidobacteria using a combination of metagenomic and cultivation approaches. Genome Biology 2019, 20, 1-6, 10.1186/s13059-019-1711-6.
- Jiri Killer; J. Kopečný; J. Mrázek; I. Koppová; J. Havlík; O. Benada; T. Kott; Bifidobacterium actinocoloniiforme sp. nov. and Bifidobacterium bohemicum sp. nov., from the bumblebee digestive tract. International Journal of Systematic and Evolutionary Microbiology 2011, 61, 1315-1321, 10.1099/ijs.0.022525-0.
- G Reuter; [COMPARATIVE STUDIES ON THE BIFIDUS FLORA IN THE FECES OF INFANTS AND ADULTS. WITH A CONTRIBUTION TO CLASSIFICATION AND NOMENCLATURE OF BIFIDUS STRAINS].. Zentralblatt fur Bakteriologie, Parasitenkunde, Infektionskrankheiten und Hygiene. 1. Abt. Medizinisch-hygienische Bakteriologie, Virusforschung und Parasitologie. Originale 1963, 191, 486-507, .
- Samanta Michelini; Monica Modesto; Gianfranco Filippini; Caterina Spiezio; Camillo Sandri; Bruno Biavati; Annamaria Pisi; Paola Mattarelli; Bifidobacterium aerophilum sp. nov., Bifidobacterium avesanii sp. nov. and Bifidobacterium ramosum sp. nov.: Three novel taxa from the faeces of cotton-top tamarin ( Saguinus oedipus L.). Systematic and Applied Microbiology 2016, 39, 229-236, 10.1016/j.syapm.2016.04.005.
- Hidehiro Toh; Yumiko Yamazaki; Kosuke Tashiro; Shinpei Kawarai; Kenshiro Oshima; Akiyo Nakano; Co Nguyen Thi Kim; Iyo Mimura; Kensuke Arakawa; Atsushi Iriki; et al. Draft Genome Sequence of Bifidobacterium aesculapii DSM 26737 T , Isolated from Feces of Baby Common Marmoset. Genome Announcements 2015, 3, e01463-15, 10.1128/genomea.01463-15.
- V. Scardovi; B. Sgorbati; Electrophoretic types of transaldolase, transketolase, and other enzymes in bifidobacteria. Antonie van Leeuwenhoek 1974, 40, 427-440, 10.1007/bf00399355.
- T Mitsuoka; [Comparative studies on bifidobacteria isolated from the alimentary tract of man and animals (including descriptions of bifidobacterium thermophilum nov. spec. and bifidobacterium pseudolongum nov. spec)].. Zentralblatt fur Bakteriologie, Parasitenkunde, Infektionskrankheiten und Hygiene. 1. Abt. Medizinisch-hygienische Bakteriologie, Virusforschung und Parasitologie. Originale 1969, 210, 52-64, .
- Liesbeth Masco; Marco Ventura; Ralf Zink; Geert Huys; Jean Swings; Polyphasic taxonomic analysis of Bifidobacterium animalis and Bifidobacterium lactis reveals relatedness at the subspecies level: reclassification of Bifidobacterium animalis as Bifidobacterium animalis subsp. animalis subsp. nov. and Bifidobacterium lactis as Bifidobacterium animalis subsp. lactis subsp. nov.. International Journal of Systematic and Evolutionary Microbiology 2004, 54, 1137-1143, 10.1099/ijs.0.03011-0.
- R. Pechar; Jiri Killer; H. Salmonová; M. Geigerová; R. Švejstil; P. Švec; I. Sedláček; V. Rada; O. Benada; Bifidobacterium apri sp. nov., a thermophilic actinobacterium isolated from the digestive tract of wild pigs (Sus scrofa). International Journal of Systematic and Evolutionary Microbiology 2017, 67, 2349-2356, 10.1099/ijsem.0.001956.
- David Laureys; Margo Cnockaert; Luc De Vuyst; Peter Vandamme; Bifidobacterium aquikefiri sp. nov., isolated from water kefir. International Journal of Systematic and Evolutionary Microbiology 2016, 66, 1281-1286, 10.1099/ijsem.0.000877.
- V Scardovi; L D Trovatelli; New species of bifid bacteria from Apis mellifica L. and Apis indica F. A contribution to the taxonomy and biochemistry of the genus Bifidobacterium.. Zentralblatt für Bakteriologie, Parasitenkunde, Infektionskrankheiten und Hygiene. Zweite Naturwissenschaftliche Abteilung: Allgemeine, Landwirtschaftliche und Technische Mikrobiologie 1969, 123, 64–88, .
- Akihito Endo; Yuka Futagawa-Endo; Peter Schumann; Rüdiger Pukall; Leon M.T. Dicks; Bifidobacterium reuteri sp. nov., Bifidobacterium callitrichos sp. nov., Bifidobacterium saguini sp. nov., Bifidobacterium stellenboschense sp. nov. and Bifidobacterium biavatii sp. nov. isolated from faeces of common marmoset (Callithrix jacchus) and red-handed tamarin (Saguinus midas). Systematic and Applied Microbiology 2012, 35, 92-97, 10.1016/j.syapm.2011.11.006.
- Jiri Killer; J. Kopečný; Jakub Mrazek; V. Rada; Oldřich Benada; I. Koppová; Jaroslav Havlík; Jakub Straka; Bifidobacterium bombi sp. nov., from the bumblebee digestive tract. International Journal of Systematic and Evolutionary Microbiology 2009, 59, 2020-2024, 10.1099/ijs.0.002915-0.
- V. Scardovi; L. D. Trovatelli; B. Biavati; G. Zani; Bifidobacterium cuniculi, Bifidobacterium choerinum, Bifidobacterium boum, and Bifidobacterium pseudocatenulatum: Four New Species and Their Deoxyribonucleic Acid Homology Relationships. International Journal of Systematic Bacteriology 1979, 29, 291-311, 10.1099/00207713-29-4-291.
- Monica Modesto; Samanta Michelini; Maria Cristina Sansosti; Carlotta De Filippo; Duccio Cavalieri; Linnea Qvirist; Thomas Andlid; Caterina Spiezio; Camillo Sandri; Stefano Pascarelli; et al. Bifidobacterium callitrichidarum sp. nov. from the faeces of the emperor tamarin (Saguinus imperator). International Journal of Systematic and Evolutionary Microbiology 2018, 68, 141-148, 10.1099/ijsem.0.002472.
- V. Scardovi; F. Crociani; Bifidobacterium catenulatum, Bifidobacterium dentium, and Bifidobacterium angulatum: Three New Species and Their Deoxyribonucleic Acid Homology Relationships. International Journal of Systematic Bacteriology 1974, 24, 6-20, 10.1099/00207713-24-1-6.
- Hidetoshi Morita; Akiyo Nakano; Hiromi Onoda; Hidehiro Toh; Kenshiro Oshima; Hideto Takami; Masaru Murakami; Shinji Fukuda; Tatsuya Takizawa; Tomomi Kuwahara; et al. Bifidobacterium kashiwanohense sp. nov., isolated from healthy infant faeces. International Journal of Systematic and Evolutionary Microbiology 2011, 61, 2610-2615, 10.1099/ijs.0.024521-0.
- Monica Modesto; Samanta Michelini; Kaihei Oki; Bruno Biavati; Koichi Watanabe; Paola Mattarelli; Bifidobacterium catulorum sp. nov., a novel taxon from the faeces of the baby common marmoset (Callithrix jacchus). International Journal of Systematic and Evolutionary Microbiology 2018, 68, 575-581, 10.1099/ijsem.0.002545.
- Jessy Praet; Ivan Meeus; Margo Cnockaert; Maarten Aerts; Guy Smagghe; Peter Vandamme; Bifidobacterium commune sp. nov. isolated from the bumble bee gut. Antonie van Leeuwenhoek 2015, 107, 1307-1313, 10.1007/s10482-015-0425-3.
- V. Delcenserie; F. Gavini; H. Beerens; Odile Tresse; C. Franssen; G. Daube; Description of a new species, Bifidobacterium crudilactis sp. nov., isolated from raw milk and raw milk cheeses. Systematic and Applied Microbiology 2007, 30, 381-389, 10.1016/j.syapm.2007.01.004.
- Samanta Michelini; Monica Modesto; Anna Maria Pisi; Gianfranco Filippini; Camillo Sandri; Caterina Spiezio; Bruno Biavati; Barbara Sgorbati; Paola Mattarelli; Bifidobacterium eulemuris sp. nov., isolated from faeces of black lemurs (Eulemur macaco). International Journal of Systematic and Evolutionary Microbiology 2016, 66, 1567-1576, 10.1099/ijsem.0.000924.
- E. Lauer; Bifidobacterium gallicum sp. nov. Isolated from Human Feces. International Journal of Systematic Bacteriology 1990, 40, 100-102, 10.1099/00207713-40-1-100.
- Samanta Michelini; Kaihei Oki; Emiko Yanokura; Yasuhisa Shimakawa; Monica Modesto; Paola Mattarelli; Bruno Biavati; Koichi Watanabe; Bifidobacterium myosotis sp. nov., Bifidobacterium tissieri sp. nov. and Bifidobacterium hapali sp. nov., isolated from faeces of baby common marmosets (Callithrix jacchus L.). International Journal of Systematic and Evolutionary Microbiology 2016, 66, 255-265, 10.1099/ijsem.0.000708.
- Monica Modesto; Koichi Watanabe; Masanori Arita; Maria Satti; Kaihei Oki; Piero Sciavilla; Claudio Patavino; Cesare Cammà; Samanta Michelini; Barbara Sgorbati; et al. Bifidobacterium jacchi sp. nov., isolated from the faeces of a baby common marmoset (Callithrix jacchus). International Journal of Systematic and Evolutionary Microbiology 2019, 69, 2477-2485, 10.1099/ijsem.0.003518.
- Monica Modesto; Samanta Michelini; Ilaria Stefanini; Camillo Sandri; Caterina Spiezio; Annamaria Pisi; Gianfranco Filippini; Bruno Biavati; Paola Mattarelli; Bifidobacterium lemurum sp. nov., from faeces of the ring-tailed lemur (Lemur catta). International Journal of Systematic and Evolutionary Microbiology 2015, 65, 1726-1734, 10.1099/ijs.0.000162.
- D. Matteuzzi; F. Crociani; O. Zani; L. D. Trovatelli; Bifidobacterium suis n. sp. : A new species of the genus Bifidobacterium isolated from pig faces. Journal of Basic Microbiology 1971, 11, 387-395, 10.1002/jobm.19710110504.
- B. Biavati; P. Mattarelli; Bifidobacterium ruminantium sp. nov. and Bifidobacterium merycicum sp. nov. from the Rumens of Cattle. International Journal of Systematic Bacteriology 1991, 41, 163-168, 10.1099/00207713-41-1-163.
- Bifidobacterium animalis (Mitsuoka) comb. nov. and the . , , , .
- Koichi Watanabe; Hiroshi Makino; Masae Sasamoto; Yuko Kudo; Junji Fujimoto; Shirchin Demberel; Bifidobacterium mongoliense sp. nov., from airag, a traditional fermented mare's milk product from Mongolia. International Journal of Systematic and Evolutionary Microbiology 2009, 59, 1535-1540, 10.1099/ijs.0.006247-0.
- Sayaka Tsuchida; Shunsuke Takahashi; Pierre Philippe Mbehang Nguema; Shiho Fujita; Maki Kitahara; Juichi Yamagiwa; Alfred Ngomanda; Moriya Ohkuma; Kazunari Ushida; Bifidobacterium moukalabense sp. nov., isolated from the faeces of wild west lowland gorilla (Gorilla gorilla gorilla). International Journal of Systematic and Evolutionary Microbiology 2014, 64, 449-455, 10.1099/ijs.0.055186-0.
- Monica Modesto; Maria Satti; Koichi Watanabe; Donatella Scarafile; Chien-Hsun Huang; Jong-Shian Liou; Tomohiko Tamura; Satomi Saito; Mizuki Watanabe; Koji Mori; et al. Phylogenetic characterization of two novel species of the genus Bifidobacterium: Bifidobacterium saimiriisciurei sp. nov. and Bifidobacterium platyrrhinorum sp. nov.. Systematic and Applied Microbiology 2020, 43, 126111, 10.1016/j.syapm.2020.126111.
- Scardovi V, Trovatelli LD, Crociani F, Sgorbati B.; Bifid bacteria in bovine rumen. New species of the genus Bifidobacterium: B. globosum n.sp. and B. ruminale n.sp. Arch. Mikrobiol. 1969, 68, 278–294, .
- Paul J. Simpson; R. Paul Ross; Gerald F. Fitzgerald; Catherine Stanton; Bifidobacterium psychraerophilum sp. nov. and Aeriscardovia aeriphila gen. nov., sp. nov., isolated from a porcine caecum. International Journal of Systematic and Evolutionary Microbiology 2004, 54, 401-406, 10.1099/ijs.0.02667-0.
- J. Watabe; Y. Benno; T. Mitsuoka; Bifidobacterium gallinarum sp. nov.: a New Species Isolated from the Ceca of Chickens. International Journal of Systematic Bacteriology 1983, 33, 127-132, 10.1099/00207713-33-2-127.
- Monica Modesto; Maria Satti; Koichi Watanabe; Edoardo Puglisi; Lorenzo Morelli; Chien-Hsun Huang; Jong-Shian Liou; Mika Miyashita; Tomohiko Tamura; Satomi Saito; et al. Characterization of Bifidobacterium species in feaces of the Egyptian fruit bat: Description of B. vespertilionis sp. nov. and B. rousetti sp. nov.. Systematic and Applied Microbiology 2019, 42, 126017, 10.1016/j.syapm.2019.126017.
- Gabriele Andrea Lugli; Christian Milani; Francesca Turroni; Sabrina Duranti; Chiara Ferrario; Alice Viappiani; Leonardo Mancabelli; Marta Mangifesta; Bernard Taminiau; Véronique Delcenserie; et al. Investigation of the Evolutionary Development of the Genus Bifidobacterium by Comparative Genomics. Applied and Environmental Microbiology 2014, 80, 6383-6394, 10.1128/aem.02004-14.
- Lesley Hoyles; Elisabeth Inganäs; Enevold Falsen; Michel Drancourt; Norbert Weiss; Anne L McCartney; Matthew D Collins; Bifidobacterium scardovii sp. nov., from human sources.. International Journal of Systematic and Evolutionary Microbiology 2002, 52, 995-999, 10.1099/00207713-52-3-995.
- Lin Zhu; Wei Li; Xiuzhu Dong; Species identification of genus Bifidobacterium based on partial HSP60 gene sequences and proposal of Bifidobacterium thermacidophilum subsp. porcinum subsp. nov.. International Journal of Systematic and Evolutionary Microbiology 2003, 53, 1619-1623, 10.1099/ijs.0.02617-0.
- X Dong; Y Xin; W Jian; X Liu; D Ling; Bifidobacterium thermacidophilum sp. nov., isolated from an anaerobic digester.. International Journal of Systematic and Evolutionary Microbiology 2000, 50, 119-125, 10.1099/00207713-50-1-119.
- Viktor P. L. Eckel; Lisa-Marie Ziegler; Rudi F. Vogel; Matthias A. Ehrmann; Bifidobacterium tibiigranuli sp. nov. isolated from homemade water kefir. International Journal of Systematic and Evolutionary Microbiology 2020, 70, 1562-1570, 10.1099/ijsem.0.003936.
- M. Okamoto; Yoshimi Benno; Kai-P Leung; Nobuko Maeda; Bifidobacterium tsurumiense sp. nov., from hamster dental plaque. International Journal of Systematic and Evolutionary Microbiology 2008, 58, 144-148, 10.1099/ijs.0.65296-0.
- Sabrina Duranti; Marta Mangifesta; Gabriele Andrea Lugli; Francesca Turroni; Rosaria Anzalone; Christian Milani; Leonardo Mancabelli; Maria Cristina Ossiprandi; Marco Ventura; Bifidobacterium vansinderenii sp. nov., isolated from faeces of emperor tamarin (Saguinus imperator). International Journal of Systematic and Evolutionary Microbiology 2017, 67, 3987-3995, 10.1099/ijsem.0.002243.
- Alessandri, G.; Milani, C.; Mancabelli, L.; Mangifesta, M.; Lugli, G.A.; Viappiani, A.; Duranti, S.; Turroni, F.; Ossiprandi, M.C.; van Sinderen, D.; et al. The impact of human-facilitated selection on the gut microbiota of domesticated mammals. FEMS Microbiol. Ecol. 2019, 95.
- Milani, C.; Mangifesta, M.; Mancabelli, L.; Lugli, G.A.; James, K.; Duranti, S.; Turroni, F.; Ferrario, C.; Ossiprandi, M.C.; van Sinderen, D.; et al. Unveiling bifidobacterial biogeography across the mammalian branch of the tree of life. ISME J. 2017, 11, 2834–2847.
- Wong, C.B.; Odamaki, T.; Xiao, J.Z. Insights into the reason of Human-Residential Bifidobacteria (HRB) being the natural inhabitants of the human gut and their potential health-promoting benefits. FEMS Microbiol. Rev. 2020, 44, 369–385.
- Bottacini, F.; Ventura, M.; van Sinderen, D.; O’Connell Motherway, M. Diversity, ecology and intestinal function of bifidobacteria. Microb. Cell. Fact. 2014, 13 (Suppl. 1), S4.
- Ruiz, L.; Delgado, S.; Ruas-Madiedo, P.; Sanchez, B.; Margolles, A. Bifidobacteria and Their Molecular Communication with the Immune System. Front. Microbiol. 2017, 8, 2345.
- Turroni, F.; Peano, C.; Pass, D.A.; Foroni, E.; Severgnini, M.; Claesson, M.J.; Kerr, C.; Hourihane, J.; Murray, D.; Fuligni, F.; et al. Diversity of bifidobacteria within the infant gut microbiota. PLoS. ONE 2012, 7, e36957.
- Palmer, C.; Bik, E.M.; DiGiulio, D.B.; Relman, D.A.; Brown, P.O. Development of the human infant intestinal microbiota. PLoS Biol. 2007, 5, e177.
- Koenig, J.E.; Spor, A.; Scalfone, N.; Fricker, A.D.; Stombaugh, J.; Knight, R.; Angenent, L.T.; Ley, R.E. Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. 1), 4578–4585.
- Avershina, E.; Lundgard, K.; Sekelja, M.; Dotterud, C.; Storro, O.; Oien, T.; Johnsen, R.; Rudi, K. Transition from infant- to adult-like gut microbiota. Environ. Microbiol. 2016, 18, 2226–2236.
- Nuriel-Ohayon, M.; Neuman, H.; Koren, O. Microbial Changes during Pregnancy, Birth, and Infancy. Front. Microbiol. 2016, 7, 1031.
- Rautava, S.; Luoto, R.; Salminen, S.; Isolauri, E. Microbial contact during pregnancy, intestinal colonization and human disease. Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 565–576.
- Christian Milani; Leonardo Mancabelli; Gabriele Andrea Lugli; Sabrina Duranti; Francesca Turroni; Chiara Ferrario; Marta Mangifesta; Alice Viappiani; Pamela Ferretti; Valentina Gorfer; et al. Exploring Vertical Transmission of Bifidobacteria from Mother to Child. Applied and Environmental Microbiology 2015, 81, 7078-7087, 10.1128/aem.02037-15.
- Sela, D.A.; Chapman, J.; Adeuya, A.; Kim, J.H.; Chen, F.; Whitehead, T.R.; Lapidus, A.; Rokhsar, D.S.; Lebrilla, C.B.; German, J.B.; et al. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc. Natl. Acad. Sci. USA 2008, 105, 18964–18969.
- Ishikawa, E.; Matsuki, T.; Kubota, H.; Makino, H.; Sakai, T.; Oishi, K.; Kushiro, A.; Fujimoto, J.; Watanabe, K.; Watanuki, M.; et al. Ethnic diversity of gut microbiota: Species characterization of Bacteroides fragilis group and genus Bifidobacterium in healthy Belgian adults, and comparison with data from Japanese subjects. J. Biosci. Bioeng. 2013, 116, 265–270.
- Odamaki, T.; Bottacini, F.; Kato, K.; Mitsuyama, E.; Yoshida, K.; Horigome, A.; Xiao, J.Z.; van Sinderen, D. Genomic diversity and distribution of Bifidobacterium longum subsp. longum across the human lifespan. Sci. Rep. 2018, 8, 85.
- Paul B. Eckburg; Elisabeth M. Bik; Charles N. Bernstein; Elizabeth Purdom; Les Dethlefsen; Michael Sargent; Steven R. Gill; Karen E. Nelson; David A. Relman; Diversity of the Human Intestinal Microbial Flora. Science 2005, 308, 1635-1638, 10.1126/science.1110591.
- Francesca Turroni; Elena Foroni; Paola Pizzetti; Vanessa Giubellini; Angela Ribbera; Paolo Merusi; Patrizio Cagnasso; Barbara Bizzarri; Gian Luigi De'angelis; Fergus Shanahan; et al. Exploring the Diversity of the Bifidobacterial Population in the Human Intestinal Tract. Applied and Environmental Microbiology 2009, 75, 1534-1545, 10.1128/aem.02216-08.
- Amy O'callaghan; Douwe Van Sinderen; Bifidobacteria and Their Role as Members of the Human Gut Microbiota. Frontiers in Microbiology 2016, 7, 925, 10.3389/fmicb.2016.00925.
- Jacob, R.A. Folate, DNA methylation, and gene expression: Factors of nature and nurture. Am. J. Clin. Nutr. 2000, 72, 903–904.
- Lucock, M. Folic acid: Nutritional biochemistry, molecular biology, and role in disease processes. Mol. Genet. Metab. 2000, 71, 121–138.
- Ganesan, B.; Weimer, B.C.; Pinzon, J.; Dao Kong, N.; Rompato, G.; Brothersen, C.; McMahon, D.J. Probiotic bacteria survive in Cheddar cheese and modify populations of other lactic acid bacteria. J. Appl. Microbiol. 2014, 116, 1642–1656.
- Heller, K.J. Probiotic bacteria in fermented foods: Product characteristics and starter organisms. Am. J. Clin. Nutr. 2001, 73, 374S–379S.
- Huang, J.S.; Bousvaros, A.; Lee, J.W.; Diaz, A.; Davidson, E.J. Efficacy of probiotic use in acute diarrhea in children: A meta-analysis. Dig. Dis. Sci. 2002, 47, 2625–2634.
- Allen, S.J.; Okoko, B.; Martinez, E.; Gregorio, G.; Dans, L.F. Probiotics for treating infectious diarrhoea. Cochrane Database Syst. Rev. 2004, CD003048.
- Saavedra, J.M.; Bauman, N.A.; Oung, I.; Perman, J.A.; Yolken, R.H. Feeding of Bifidobacterium bifidum and Streptococcus thermophilus to infants in hospital for prevention of diarrhoea and shedding of rotavirus. Lancet 1994, 344, 1046–1049.
