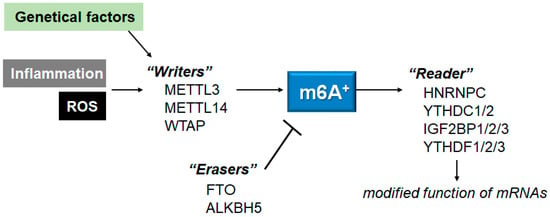
Changes in epitranscriptome with N6-methyladenine (m6A) modification could be involved in the development of multiple diseases, which might be a prevalent modification of messenger RNAs (mRNAs) in eukaryotes. The m6A modification might be performed through the action of methyltransferases, demethylases, and methylation-binding proteins. Importantly, the m6A methylation may be associated with various neurological disorders including Alzheimer’s disease (AD), Parkinson’s disease (PD), depression, aging-related diseases, and/or aging itself. In addition, the m6A methylation might functionally regulate the eukaryotic transcriptome by influencing the splicing, export, subcellular localization, translation, stability, and decay of mRNAs.
- N6-methyladenine
- reactive oxygen species
- RNA-binding protein
1. Introduction
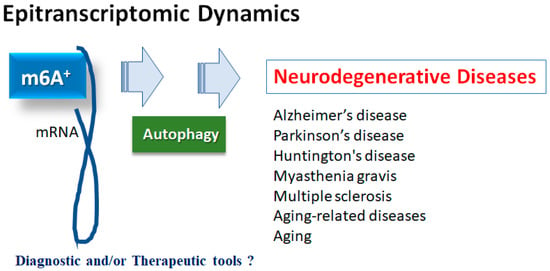
2. m6A Methylation of mRNAs in Neurodegenerative Diseases
2.1. Alzheimer’s Disease
2.2. Parkinson’s Disease
2.3. Neurodegenerative Disorders and Aging
3. ROS, Inflammation and m6A mRNAs
This entry is adapted from the peer-reviewed paper 10.3390/neurolint15030062
References
- Wang, M.; Pan, W.; Xu, Y.; Zhang, J.; Wan, J.; Jiang, H. Microglia-mediated neuroinflammation: A potential target for the treatment of cardiovascular diseases. J. Inflamm. Res. 2022, 15, 3083–3094.
- Martinez-Martin, P.; Rodriguez-Blazquez, C.; Forjaz, M.J. Quality of life and burden in caregivers for patients with Parkinson’s disease: Concepts, assessment and related factors. Expert Rev. Pharmacoecon. Outcomes Res. 2012, 12, 221–230.
- Brodaty, H.; Breteler, M.M.; DeKosky, S.T.; Dorenlot, P.; Fratiglioni, L.; Hock, C.; Kenigsberg, P.A.; Scheltens, P.; De Strooper, B. The world of dementia beyond 2020. J. Am. Geriatr. Soc. 2011, 59, 923–927.
- Do, M.D.; Tran, T.N.; Luong, A.B.; Le, L.H.G.; Van Le, T.; Le, K.T.; Van Vo, N.T.; Le, T.N.N.; Vu, H.A.; Mai, T.P.; et al. Clinical and genetic analysis of Vietnamese patients diagnosed with early-onset Parkinson’s disease. Brain Behav. 2023, 13, e2950.
- Naseri, N.; Sharma, M.; Velinov, M. Autosomal dominant neuronal ceroid lipofuscinosis: Clinical features and molecular basis. Clin. Genet. 2021, 99, 111–118.
- Cantara, W.A.; Crain, P.F.; Rozenski, J.; McCloskey, J.A.; Harris, K.A.; Zhang, X.; Vendeix, F.A.; Fabris, D.; Agris, P.F. The RNA modification database, RNAMDB: 2011 update. Nucleic Acids Res. 2010, 39 (Suppl. S1), D195–D201.
- Li, Y.; Xiao, J.; Bai, J.; Tian, Y.; Qu, Y.; Chen, X.; Wang, Q.; Li, X.; Zhang, Y.; Xu, J. Molecular characterization and clinical relevance of m6A regulators across 33 cancer types. J. Mol. Cancer 2019, 18, 137.
- Ren, J.; Li, Y.; Wuermanbieke, S.; Hu, S.; Huang, G. N6-methyladenosine (m6A) methyltransferase METTL3-mediated LINC00680 accelerates osteoarthritis through m6A/SIRT1 manner. Cell Death Discov. 2022, 8, 240.
- Weng, Y.-L.; Wang, X.; An, R.; Cassin, J.; Vissers, C.; Liu, Y.; Liu, Y.; Xu, T.; Wang, X.; Wong, S.Z.H.; et al. Epitranscriptomic m6A regulation of axon regeneration in the adult mammalian nervous system. Neuron 2018, 97, 313–325.e316.
- Widagdo, J.; Anggono, V. The m6A-epitranscriptomic signature in neurobiology: From neurodevelopment to brain plasticity. J. Neurochem. 2018, 147, 137–152.
- Livneh, I.; Moshitch-Moshkovitz, S.; Amariglio, N.; Rechavi, G.; Dominissini, D. The m6A epitranscriptome: Transcriptome plasticity in brain development and function. Nat. Rev. Neurosci. 2020, 21, 36–51.
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485, 201–206.
- Lu, S.; Ding, X.; Wang, Y.; Hu, X.; Sun, T.; Wei, M.; Wang, X.; Wu, H. The relationship between the network of non-coding RNAs-molecular targets and N6-methyladenosine modification in colorectal cancer. Front. Cell Dev. Biol. 2021, 9, 772542.
- Ma, S.; Chen, C.; Ji, X.; Liu, J.; Zhou, Q.; Wang, G.; Yuan, W.; Kan, Q.; Sun, Z. The interplay between m6A RNA methylation and noncoding RNA in cancer. J. Hematol. Oncol. 2019, 12, 121.
- Zhao, B.S.; Roundtree, I.A.; He, C. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 2017, 18, 31–42.
- Zhang, H.; Shi, X.; Huang, T.; Zhao, X.; Chen, W.; Gu, N.; Zhang, R. Dynamic landscape and evolution of m6A methylation in human. Nucleic Acids Res. 2020, 48, 6251–6264.
- Wang, W.; Dong, D.; Yu, P.; Chen, T.; Gao, R.; Wei, J.; Mo, Z.; Zhou, H.; Yang, Q.; Yue, C.; et al. Prognostic model based on m6A -associated lncRNAs in esophageal cancer. Front. Endocrinol. 2022, 13, 947708.
- Zhou, H.; Wang, B.; Sun, H.; Xu, X.; Wang, Y. Epigenetic regulations in neural stem cells and neurological diseases. Stem Cells Int. 2018, 2018, 6087143.
- Li, J.; Yang, X.; Qi, Z.; Sang, Y.; Liu, Y.; Xu, B.; Liu, W.; Xu, Z.; Deng, Y. The role of mRNA m6A methylation in the nervous system. Cell Biosci. 2019, 9, 66.
- Liu, S.; Li, Q.; Chen, K.; Zhang, Q.; Li, G.; Zhuo, L.; Zhai, B.; Sui, X.; Hu, X.; Xie, T. The emerging molecular mechanism of m6A modulators in tumorigenesis and cancer progression. Biomed. Pharmacother. 2020, 127, 110098.
- Song, H.; Feng, X.; Zhang, H.; Luo, Y.; Huang, J.; Lin, M.; Jin, J.; Ding, X.; Wu, S.; Huang, H.; et al. METTL3 and ALKBH5 oppositely regulate m6A modification of TFEB mRNA, which dictates the fate of hypoxia/reoxygenation-treated cardiomyocytes. Autophagy 2019, 15, 1419–1437.
- Wang, X.; Wu, R.; Liu, Y.; Zhao, Y.; Bi, Z.; Yao, Y.; Liu, Q.; Shi, H.; Wang, F.; Wang, Y. m6A mRNA methylation controls autophagy and adipogenesis by targeting Atg5 and Atg7. Autophagy 2020, 16, 1221–1235.
- Liu, S.; Li, Q.; Li, G.; Zhang, Q.; Zhuo, L.; Han, X.; Zhang, M.; Chen, X.; Pan, T.; Yan, L.; et al. The mechanism of m6A methyltransferase METTL3-mediated autophagy in reversing gefitinib resistance in NSCLC cells by β-elemene. Cell Death Dis. 2020, 11, 969.
- Li, G.; Luo, R.; Zhang, W.; He, S.; Wang, B.; Liang, H.; Song, Y.; Ke, W.; Shi, Y.; Feng, X.; et al. m6A hypomethylation of DNMT3B regulated by ALKBH5 promotes intervertebral disc degeneration via E4F1 deficiency. Clin. Transl. Med. 2022, 12, e765.
- Han, M.; Liu, Z.; Xu, Y.; Liu, X.; Wang, D.; Li, F.; Wang, Y.; Bi, J. Abnormality of m6A mRNA methylation is involved in Alzheimer’s disease. J. Front. Neurosci. 2020, 14, 98.
- Pinheiro, L.; Faustino, C. Therapeutic strategies targeting amyloid-β in Alzheimer’s disease. Curr. Alzheimer Res. 2019, 16, 418–452.
- Shafik, A.M.; Zhang, F.; Guo, Z.; Dai, Q.; Pajdzik, K.; Li, Y.; Kang, Y.; Yao, B.; Wu, H.; He, C.; et al. N6-methyladenosine dynamics in neurodevelopment and aging, and its potential role in Alzheimer’s disease. Genome Biol. 2021, 22, 17.
- Shao, N.; Ye, T.; Xuan, W.; Zhang, M.; Chen, Q.; Liu, J.; Zhou, P.; Song, H.; Cai, B. The effects of N 6-methyladenosine RNA methylation on the nervous system. Mol. Cell Biochem. 2023, 1–13.
- Tang, Z.; Cao, J.; Yao, J.; Fan, X.; Zhao, J.; Zhao, M.; Duan, Q.; Han, B.; Duan, S. KDM1A-mediated upregulation of METTL3 ameliorates Alzheimer’s disease via enhancing autophagic clearance of p-Tau through m6A-dependent regulation of STUB1. Free Radic. Biol. Med. 2023, 195, 343–358.
- Hao, X.; Li, Y.; Huang, G.; Zeng, Y. Role of the N6-methyladenosine regulatory factor in reducing the risk of cardiovascular disease: Subtype diagnosis following aerobic exercise-assisted weight loss. Am. J. Transl. Res. 2022, 14, 5363.
- Teng, Y.; Liu, Z.; Chen, X.; Liu, Y.; Geng, F.; Le, W.; Jiang, H.; Yang, L. Conditional deficiency of m6A methyltransferase Mettl14 in substantia nigra alters dopaminergic neuron function. J. Cell. Mol. Med. 2021, 25, 8567–8572.
- Chen, X.; Yu, C.; Guo, M.; Zheng, X.; Ali, S.; Huang, H.; Zhang, L.; Wang, S.; Huang, Y.; Qie, S.; et al. Down-regulation of m6A mRNA methylation is involved in dopaminergic neuronal death. ACS Chem. Neurosci. 2019, 10, 2355–2363.
- Qi, Z.; Wang, S.; Li, J.; Wen, Y.; Cui, R.; Zhang, K.; Liu, Y.; Yang, X.; Zhang, L.; Xu, B.; et al. Protective role of mRNA demethylase FTO on axon guidance molecules of nigro-striatal projection system in manganese-induced parkinsonism. J. Hazard. Mater. 2022, 426, 128099.
- Ondo, K.; Isono, M.; Nakano, M.; Hashiba, S.; Fukami, T.; Nakajima, M. The N6-methyladenosine modification posttranscriptionally regulates hepatic UGT2B7 expression. Biochem. Pharmacol. 2021, 189, 114402.
- Foroud, T.; Siemers, E.; Kleindorfer, D.; Bill, D.J.; Hodes, M.; Norton, J.A.; Conneally, P.M.; Christian, J.C. Cognitive scores in carriers of Huntington’s disease gene compared to noncarriers. Ann. Neurol. 1995, 37, 657–664.
- Harris, K.L.; Armstrong, M.; Swain, R.; Erzinclioglu, S.; Das, T.; Burgess, N.; Barker, R.A.; Mason, S.L. Huntington’s disease patients display progressive deficits in hippocampal-dependent cognition during a task of spatial memory. Cortex 2019, 119, 417–427.
- Pupak, A.; Singh, A.; Sancho-Balsells, A.; Alcalá-Vida, R.; Espina, M.; Giralt, A.; Martí, E.; Ørom, U.A.V.; Ginés, S.; Brito, V. Altered m6A RNA methylation contributes to hippocampal memory deficits in Huntington’s disease mice. Cell. Mol. Life Sci. 2022, 79, 416.
- Gomez, A.M.; Van Den Broeck, J.; Vrolix, K.; Janssen, S.P.; Lemmens, M.A.; Van Der Esch, E.; Duimel, H.; Frederik, P.; Molenaar, P.C.; Martínez-Martínez, P.; et al. Antibody effector mechanisms in myasthenia gravis—Pathogenesis at the neuromuscular junction. Autoimmunity 2010, 43, 353–370.
- Li, S.; Liu, H.; Ruan, Z.; Guo, R.; Sun, C.; Tang, Y.; Huang, X.; Gao, T.; Hao, S.; Li, H.; et al. Landscape analysis of m6A modification regulators related biological functions and immune characteristics in myasthenia gravis. J. Transl. Med. 2023, 21, 166.
- Thompson, A.J.; Banwell, B.L.; Barkhof, F.; Carroll, W.M.; Coetzee, T.; Comi, G.; Correale, J.; Fazekas, F.; Filippi, M.; Freedman, M.S.; et al. Diagnosis of multiple sclerosis: 2017 revisions of the McDonald criteria. Lancet Neurol. 2018, 17, 162–173.
- Dalmau, J. Diagnostic and treatment challenges, a new section in N2. Neurol. Neuroimmunol. Neuroinflamm. 2018, 5, e511.
- Mo, X.-B.; Lei, S.-F.; Qian, Q.-Y.; Guo, Y.-F.; Zhang, Y.-H.; Zhang, H. Integrative analysis revealed potential causal genetic and epigenetic factors for multiple sclerosis. J. Neurol. 2019, 266, 2699–2709.
- Castro-Hernández, R.; Berulava, T.; Metelova, M.; Epple, R.; Peña Centeno, T.; Richter, J.; Kaurani, L.; Pradhan, R.; Sakib, M.S.; Burkhardt, S.; et al. Conserved reduction of m6A RNA modifications during aging and neurodegeneration is linked to changes in synaptic transcripts. Proc. Natl. Acad. Sci. USA 2023, 120, e2204933120.
- Su, X.; Shen, Y.; Jin, Y.; Kim, I.-M.; Weintraub, N.L.; Tang, Y. Aging-associated differences in epitranscriptomic m6A regulation in response to acute cardiac ischemia/reperfusion injury in female mice. Front. Pharmacol. 2021, 12, 654316.
- Chen, Y.-S.; Ouyang, X.-P.; Yu, X.-H.; Novák, P.; Zhou, L.; He, P.-P.; Yin, K. N6-Adenosine methylation (m6A) RNA modification: An emerging role in cardiovascular diseases. J. Cardiovasc. Transl. Res. 2021, 14, 857–872.
- Shi, H.; Wei, J.; He, C. Where, when, and how: Context-dependent functions of RNA methylation writers, readers, and erasers. Mol. Cell 2019, 74, 640–650.
- Luo, J.; Xu, T.; Sun, K. N6-methyladenosine RNA modification in inflammation: Roles, mechanisms, and applications. Front. Cell Dev. Biol. 2021, 9, 670711.
- Mapperley, C.; van de Lagemaat, L.N.; Lawson, H.; Tavosanis, A.; Paris, J.; Campos, J.; Wotherspoon, D.; Durko, J.; Sarapuu, A.; Choe, J.; et al. The mRNA m6A reader YTHDF2 suppresses proinflammatory pathways and sustains hematopoietic stem cell function. J. Exp. Med. 2020, 218, e20200829.
- Gong, C.; Wu, J.; Li, H.; Luo, C.; Ji, G.; Guan, X.; Liu, J.; Wang, M. METTL3 achieves lipopolysaccharide-induced myocardial injury via m6A -dependent stabilization of Myh3 mRNA. Biochim. Biophys. Acta Mol. Cell Res. 2023, 1870, 119503.
- Feng, Z.; Li, Q.; Meng, R.; Yi, B.; Xu, Q. METTL 3 regulates alternative splicing of MyD88 upon the lipopolysaccharide-induced inflammatory response in human dental pulp cells. J. Cell. Mol. Med. 2018, 22, 2558–2568.
- Wang, A.; Jin, C.; Wang, Y.; Yu, J.; Wang, R.; Tian, X. FTO promotes the progression of cervical cancer by regulating the N6-methyladenosine modification of ZEB1 and Myc. Mol. Carcinog. 2023, 62, 1228–1237.
- Luo, J.; Wang, F.; Sun, F.; Yue, T.; Zhou, Q.; Yang, C.; Rong, S.; Yang, P.; Xiong, F.; Yu, Q.; et al. Targeted inhibition of FTO demethylase protects mice against LPS-induced septic shock by suppressing NLRP3 inflammasome. Front. Immunol. 2021, 12, 663295.
- Yue, Y.; Liu, J.; He, C. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 2015, 29, 1343–1355.
- Roundtree, I.A.; Evans, M.E.; Pan, T.; He, C. Dynamic RNA modifications in gene expression regulation. Cell 2017, 169, 1187–1200.
- Deng, Y.; Zhu, H.; Xiao, L.; Liu, C.; Liu, Y.-L.; Gao, W. Identification of the function and mechanism of m6A reader IGF2BP2 in Alzheimer’s disease. Aging 2021, 13, 24086.
- Yang, Y.; Hsu, P.J.; Chen, Y.-S.; Yang, Y.-G. Dynamic transcriptomic m6A decoration: Writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018, 28, 616–624.
- Guo, F.; Zhang, Y.; Ma, J.; Yu, Y.; Wang, Q.; Gao, P.; Wang, L.; Xu, Z.; Wei, X.; Jing, M. m6A mRNA Methylation Was Associated With Gene Expression and Lipid Metabolism in Liver of Broilers Under Lipopolysaccharide Stimulation. Front. Genet. 2022, 13, 818357.
- Qi, L.; Wang, Y.; Hu, H.; Li, P.; Hu, H.; Li, Y.; Wang, K.; Zhao, Y.; Feng, M.; Lyu, H.; et al. m6A methyltransferase METTL3 participated in sympathetic neural remodeling post-MI via the TRAF6/NF-κB pathway and ROS production. J. Mol. Cell. Cardiol. 2022, 170, 87–99.
- Zhong, X.; Yu, J.; Frazier, K.; Weng, X.; Li, Y.; Cham, C.M.; Dolan, K.; Zhu, X.; Hubert, N.; Tao, Y.; et al. Circadian clock regulation of hepatic lipid metabolism by modulation of m6A mRNA methylation. Cell Rep. 2018, 25, 1816–1828.e4.
- Han, Y.-C.; Xie, H.-Z.; Lu, B.; Xiang, R.-L.; Zhang, H.-P.; Li, J.-Y.; Zhang, S.-Y. Lipopolysaccharide alters the m6A epitranscriptomic tagging of RNAs in cardiac tissue. Front. Mol. Biosci. 2021, 8, 670160.
- Xu, Q.; Wang, Y.; Chen, Z.; Yue, Y.; Huang, H.; Wu, B.; Liu, Y.; Zhou, D.-X.; Zhao, Y. ROS-stimulated protein lysine acetylation is required for crown root development in rice. J. Adv. Res. 2023, 48, 33–46.
- Wu, Q.; Ni, X. ROS-mediated DNA methylation pattern alterations in carcinogenesis. Curr. Drug Targets 2015, 16, 13–19.
- Xu, M.; Zhuo, R.; Tao, S.; Liang, Y.; Liu, C.; Liu, Q.; Wang, T.; Zhong, X. m6A RNA Methylation Mediates NOD1/NF-kB Signaling Activation in the Liver of Piglets Challenged with Lipopolysaccharide. Antioxidants 2022, 11, 1954.
