The triamine spermidine is a key metabolite of the polyamine pathway. It plays a crucial role in many infectious diseases caused by viral or parasitic infections. Spermidine and its metabolizing enzymes, i.e., spermidine/spermine-N1-acetyltransferase, spermine oxidase, acetyl polyamine oxidase, and deoxyhypusine synthase, fulfill common functions during infection in parasitic protozoa and viruses which are obligate, intracellular parasites. The competition for this important polyamine between the infected host cell and the pathogen determines the severity of infection in disabling human parasites and pathogenic viruses.
1. The Biosynthesis and Metabolism of the Triamine Spermidine in the Human Host Cell and Its Regulation
The triamine spermidine is a biogenic basic polyamine that is essential for the human host cell. In eukaryotes, polyamines can be formed either from the amino acids ornithine (Orn) or arginine (Arg). However, the biosynthesis of PAs exclusively starts from Arg in mammalian cells (Figure 1) with arginase (Arg1) catalyzing the reaction from Arg to Orn.
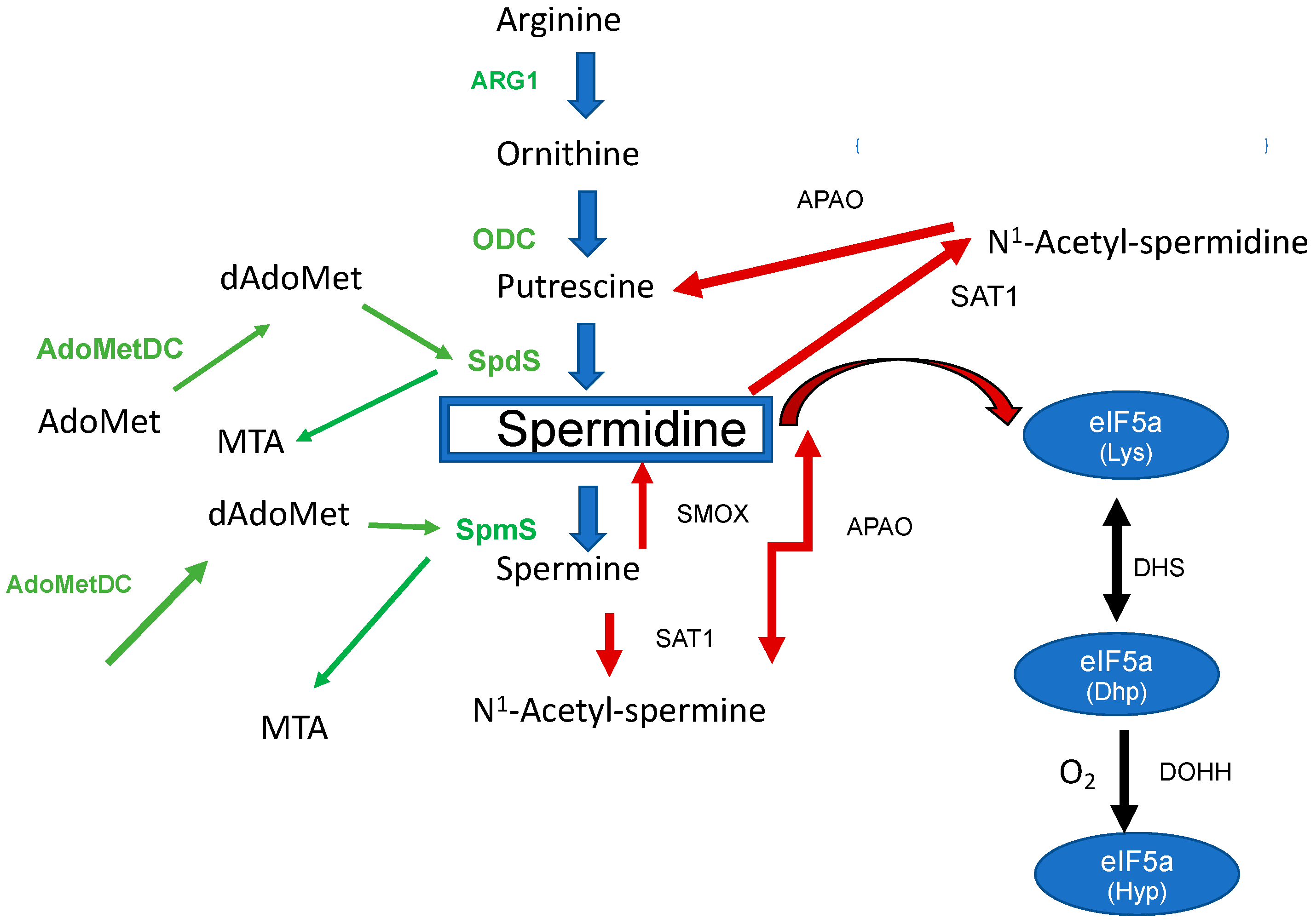
Figure 1. The biosynthesis of spermidine and its metabolism in the mammalian host. Enzymes involved in the biosynthesis are labeled in green: Arg1 (arginase), ODC, SpdS, SpmS, AdoMetDC (S-Adenosylmethionine decarboxylase), dAdoMet (decarboxylated S-Adenosylmethionine), and Methylthioadenosine (MTA). Spermidine metabolizing enzymes are marked with red arrows: PAOX, SAT1, and Spermine oxidase (SMOX). The right part of the figure shows the biosynthesis to hypusine: DHS (deoxyhypusine synthase) and DOHH (deoxyhypusine hydroxylase).
The second step of the polyamine pathway
[1] (
Figure 1) is initiated by the decarboxylation of Orn to Put by ornithine decarboxylase (ODC). Within the next step, spermidine synthase (SpdS) transfers an aminopropyl moiety from decarboxylated S-adenosylmethionine (dSAM) to Put which results in the formation of spermidine (Spd). Spermine (Spm) is formed from Spd under the catalysis of spermine synthase (SpmS). In this reaction an aminopropyl moiety is transferred from dSAM to form Spm. Collectively, there are two aminopropyltransferases which catalyze the addition of aminopropyl groups sequentially.
2. The Impact of Spermidine and Its Metabolites in Infection with Human Pathogenic Viruses
2.1. Dysregulation of Spermidine Metabolism Determines SARS-CoV-2 Infection
The role of Spd and its metabolites in mammalian viral propagation is diverse. Their main functions during infection can be attributed to viral entry, transcription, replication, and virion packaging. The impact of Spd in infections has been recently demonstrated in an in-depth metabolic study for SARS-CoV-2
[2] during the pandemic from 2020 to 2022. SARS-CoV-2 is an RNA virus that belongs to the coronavirus family. According to the current knowledge, it is zoonotically transmitted to humans, similar to the SARS virus. For replication in the human host, it needs binding of its spike proteins to the Angiotensin converting enzyme receptor 2 (ACE). The entrance of SARS-CoV-2 into the human host cell is only possible with the host-specific protease TMPRSS2. SARS-CoV-2 is mainly transmitted by droplet infection and can enter the human host via the respiratory tract, the eyes, the nose, or the mouth. Primarily, the respiratory tract is affected but other organs can also be affected. Infected people with mild symptoms, or even without, can transmit the disease
[3]. Upon infection, SARS-CoV-2 causes a dysregulation of important key metabolites in the human host cell. The virus reduces autophagy which causes degradation of macromolecules into their monomers, such as amino acids and fatty acids, after fusion with acidic lysosomes. Spd plays a key role in these processes since it is an inducer of autophagy
[4] (
Figure 2): (i) The acetylated forms of Spd promote the export of polyamines and reduce viral propagation
[5]; (ii) moreover, Spd as a building block in hypusine formation of eIF5A, activates translation of the transcription factor TFBE which induces a set of proteins involved in autophagy
[6]; (iii) interestingly,
SAT1 is transcriptionally upregulated during SARS-CoV-2 infection in vitro and in vivo. This result was confirmed by metabolome analysis which revealed an increase in putrescine levels. This shift to putrescine reduces the activity of hypusinated eIF5A and the translation of TFBE m-RNA
[2].
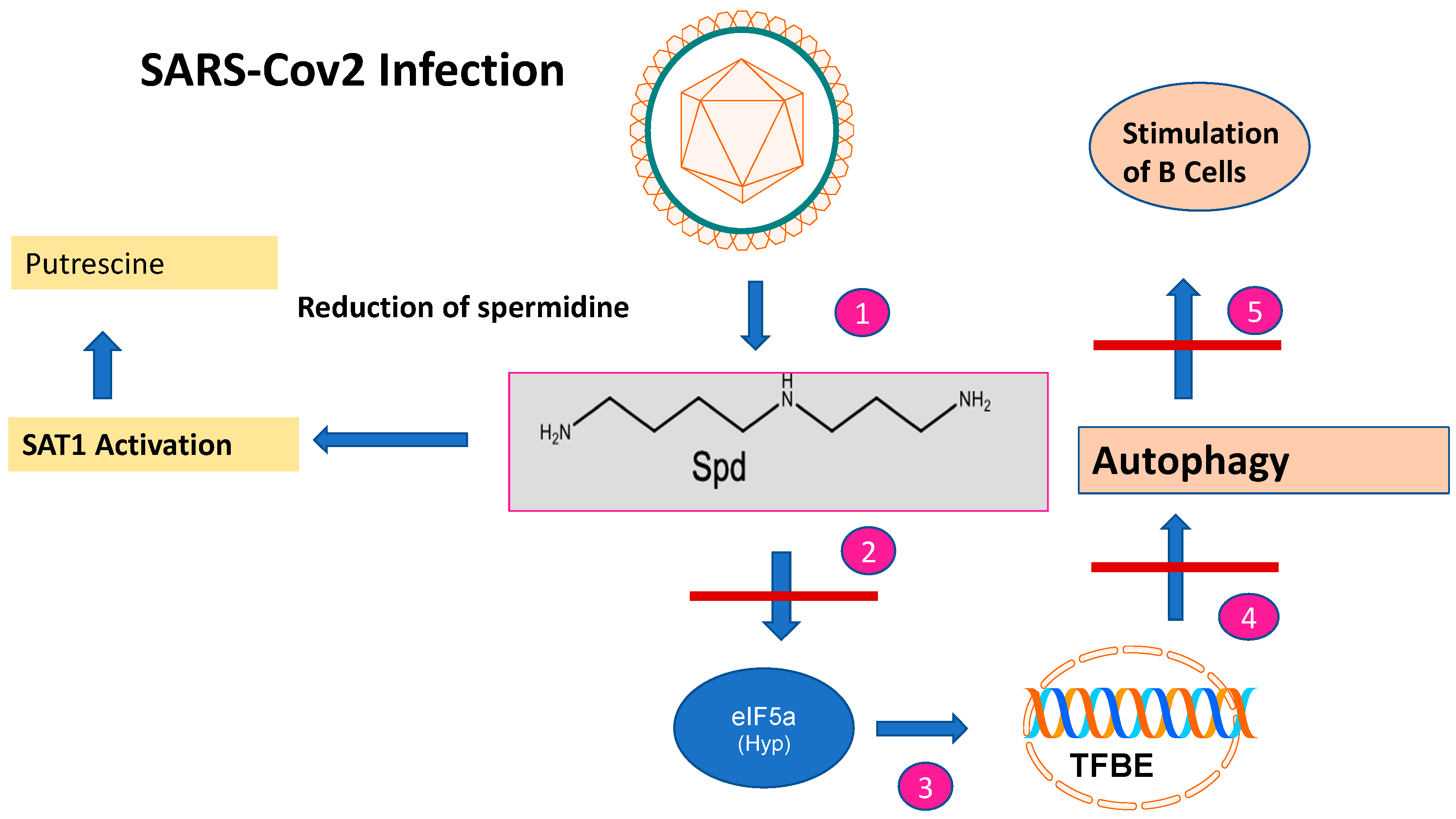
Figure 2. The impact of SARS-CoV-2 infection on spermidine metabolism. After SARS-CoV-2 infection, the spermidine concentration decreases (step 1). This is caused by SAT1 activation and a metabolic shift to putrescine. The decreased spermidine concentration affects EIF5A leading to reduced EIF-5A formation (step 2) and subsequent translation of the TFBE transcription factor (step 3). This results in reduced transcription of genes involved in autophagy (step 4) and stimulation of B cells involved in immune responses (step 5).
2.2. The Role of Hypusinated EIF-5A and PAs in Ebola Propagation
Ebola virus (EBOV) is a nonsegmented negative-sense RNA virus in the Filoviridae family. It causes Ebola virus disease (EBOVD). EBOVD starts from a single case of probable zoonotic transmission; while in the next step, human to human transmission by infected body fluids follows
[7]. Ebola virus infections belong to the deadliest viruses, with fatality rates between 40% and 90%.
In 2019, the first vaccine against EBOVD was approved by the European Medicines Agency
[8]. Apart from remdesivir
[9], two monoclonal antibodies targeting the envelope glycoprotein showed promising results
[10]. The Ebola genome has a small size of 19 kb, expressing seven structural proteins and seven non-structural proteins. Seven genes encode the following proteins:
NP nucleoprotein, VP35 polymerase cofactor, VP40 matrix protein, GP glycoprotein and secreted glycoproteins, VP30 transcriptional activator, VP24 RNA complex-associated protein, and L large protein. These viral proteins are responsible for the transcription and replication of the EBOV genome before the assembly and egress of new viral particles. Polyamines from the host cell are also essential for the replication of EBOV
[11]. Most viruses lack an intact polyamine pathway, barring one exception, i.e., PBC-V1 virus
[12]. This virus encodes a complete polyamine pathway in its ds DNA genome and infects a chlorella-like green alga. Therefore, in the case of EBOV, the question arises whether cellular PAs from the human host are necessary at the level of translation such as spermidine in modified eIF5A or in multiple stages of gene expression due to their diverse cellular functions (
Figure 3).
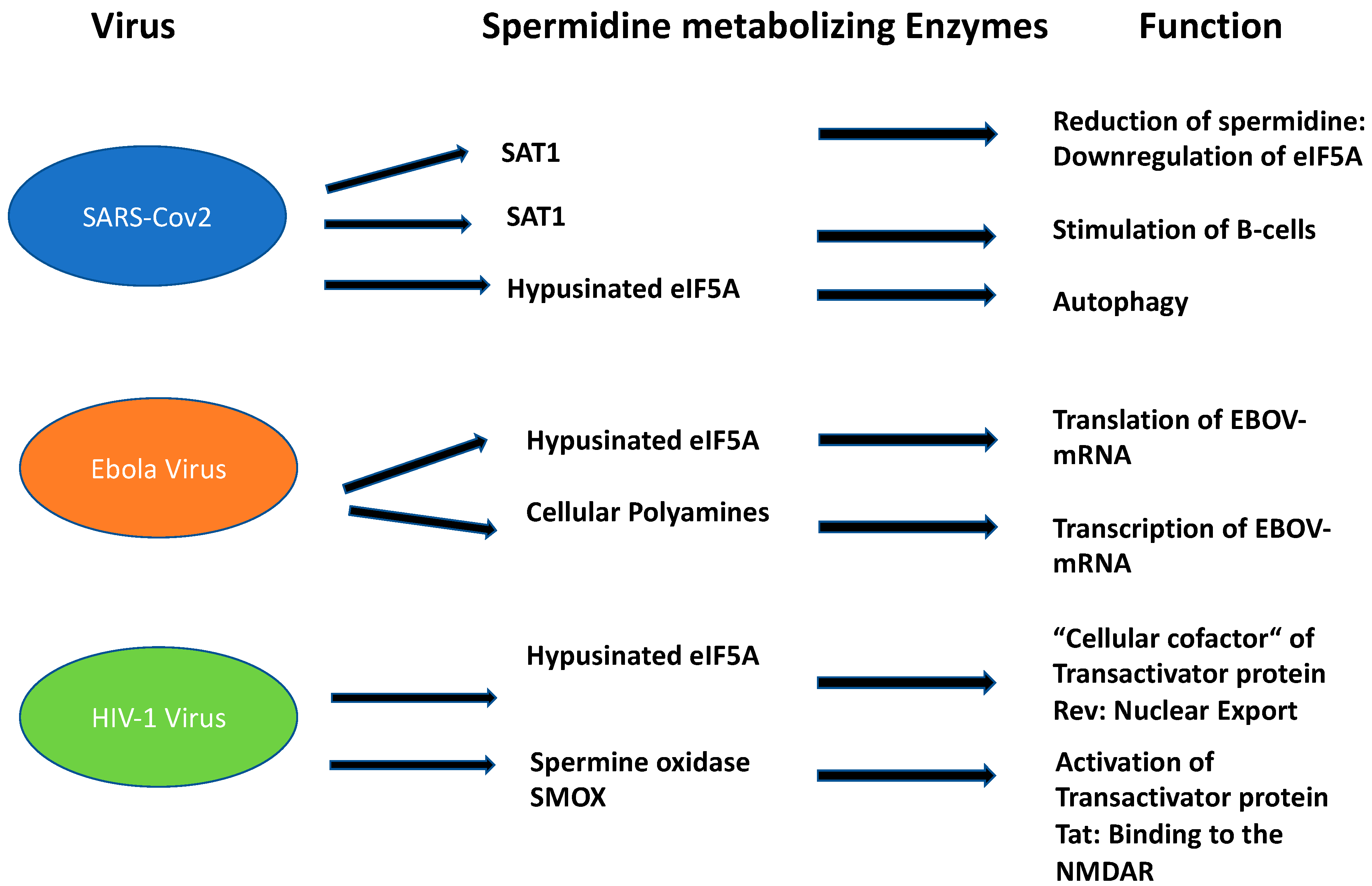
Figure 3. Function of spermidine and its metabolizing enzymes in the three most important disabling human viruses.
2.3. Hypusinated eIF5A Is a Cofactor of HIV-1 Rev Regulatory Protein
More than 76 million people worldwide are infected with the human immunodeficiency virus (HIV) which causes acquired immune deficiency syndrome (AIDS). There are now 37 million people living with the infection
[13] due to a highly effective antiretroviral therapy (HAART). Human immunodeficiency is caused by a collection of viruses, with human immunodeficiency virus 1 (HIV-1) being the most prevalent and HIV-2 being less pathogenic. HIV-1 is an RNA virus of the genus Lentivirus which belongs to the family of
Retroviridae. The disease leads to progressive loss of CDC4
+ cells, causing infectious and immune abnormalities and oncological complications. Meanwhile, antiretroviral drugs are highly efficient for patients who can access and adhere to them, thus leading to durable and probable long-life suppression of viral replication. HIV is a retrovirus that integrates its DNA into the human genome. The main receptor for HIV virus is the CD4 receptor together with two chemokine receptors, CCR5 and CXCR4. These receptors are expressed on T lymphocytes, macrophages, and monocytes. After fusion and uncoating, the RNA is reverse transcribed into DNA. Subsequently, HIV DNA is transcribed to viral mRNA, after import in the nucleus. These HIV mRNAs are then exported to the cytoplasm where translation occurs to make viral proteins and eventually mature virions. Each step of the HIV life cycle, i.e., HIV entry, reverse transcription, integration, and protein maturation, can be targeted by antiretroviral drugs. EIF5A plays an important role in HIV-1 replication since it is a “cellular cofactor” of the HIV-1 trans-activator protein Rev (
Figure 3)
[14].
3. Spermidine as a Hallmark in the Apicomplexan Parasite P. falciparum
Apicomplexan parasites represent a phylum with an apical complex; a conical structured organelle at the apical end of the cell that enables the parasite to penetrate the host by micronemes, rhoptries, conoid, and polar rings
[15]. The apicomplexan parasites of medical importance include
Plasmodium spp. (causing malaria),
Cryptosporidium parvum (causing Cryptosporidiosis),
Babesia sp. (causing Babesiosis), and
Toxoplasma gondii (causing Toxoplasmosis). Most of the apicomplexan parasites are sporozoans but some of them also produce oocytes.
Almost 50 years after the establishment of the WHO Global Malaria Programme, malaria remains a major global health problem with an estimated 627,000 deaths in 2020, according to a WHO report published in 2021
[16]. Although the number of deaths decreased between 2000 and 2019, SARS-CoV-2 refueled the rate of infection. Moreover, the death rate of children under the age of five remained at 72%. The development of the RTS,S vaccine against the parasite’s circumsporozoite protein was a milestone although it showed only a short-lived immune protection in phase III clinical trials in different African countries
[17]. Therefore, next-generation vaccines with prolonged immune protection are necessary and are beyond the horizon
[18]. Eradication of malaria is a major problem, since its causative agent,
P. falciparum, has a complex life cycle and can remain latent within the human host
[19]. Conclusively, eradication of malaria will only be successful with a combined therapy of vaccination and small molecules due to the developing resistance against registered drugs.
Plasmodium ssp. perform de novo polyamine biosynthesis but also use a salvage pathway for the uptake of Put and Spd
[20][21] in the infected erythrocyte. The first reports about highly specific Put and Spd carriers demonstrated novel transporters with different kinetic parameters compared to transporters in the uninfected erythrocyte
[20]. Later, radioisotope flux techniques proved the uptake of Put and Spd in a temperature dependent process from the infected erythrocyte
[21].
Plasmodium is the only apicomplexan parasite which contains a full set of key enzymes involved in the biosynthesis of spermidine (the core pathway), i.e., ODC, AdoMetDC, and SpdS (
Figure 4), and a conserved pathway leading to the posttranslational modification of hypusine in eIF5A.
Figure 4. De novo biosynthesis of polyamines (PAs) in
Plasmodium falciparum.
Plasmodium has a core PA pathway with ornithine decarboxylase (ODC), S-adenosylmethionine decarboxylase (AdoMetDC), and spermidine synthase (SpdS). A bifunctional AdoMetDC and a SpS producing Spm are peculiar for this pathway in
Plasmodium. The parasite is also able to use Put and Spd from the salvage pathway of the infected host red blood cell (RBC). Hypusine biosynthesis is highly conserved. Deoxhypusine synthase (DHS) catalyzes the transfer of the aminopropyl moiety to lysine 50 in the precursor protein and deoxyhypusine hydroxylase (DOHH) introduces the hydroxyl group to carbon 9 in the side chain.
4. Spermidine Biosynthesis and Its Metabolizing Pathways in the Trypanosomatids Are Unique
The trypanosomatids cause a variety of insect-born pathogenic, infectious, human diseases such as human African trypanosomiasis (
T. brucei), Chagas disease (
T. cruzi), and Leishmaniasis (
Leishmania species). According to WHO reports
[22], an estimated 18 million people are infected worldwide. In trypanosomatids, a core PA pathway and a trypanothione pathway exist as unique metabolic pathways
[23]. They both utilize spermidine as a key metabolite (
Figure 5). Most of the enzymes of both pathways are essential for survival, growth, and infectivity. One peculiar feature of the core PA pathway is the occurrence of pseudoenzymes that evolved from enzymes into regulators
[24]. In this context, AdoMetDC and DHS are outstanding examples (
Figure 5). However, trypanosomatids lack catabolizing enzymes such as SMOX, SSAT, and APAO although some catabolites produced by these enzymes appear in the parasite.
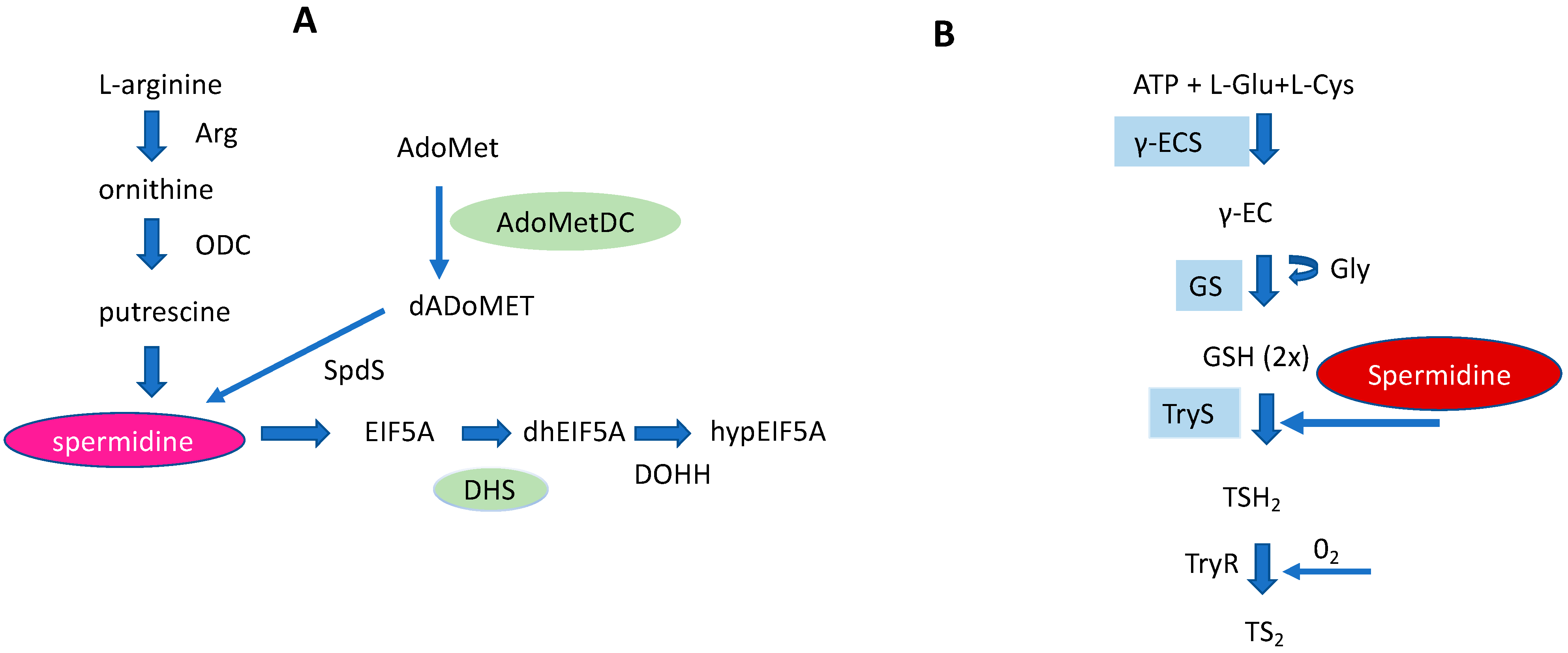
Figure 5. Schematic representation of a core PA pathway (A), and a Trypanothione pathway (B) in Trypanosomatids. In both pathways spermidine represents an important key metabolite. Pseudoenzymes, i.e., DHS and AdoMetDC of a core PA pathway are marked in green. The right part of the figure: Cys (cysteine) and Glu (glutamate) are covalently linked by γECS (gamma-glutamylcysteine synthetase) to form γEC (gamma-glutamylcysteine). Gly (Glycine) is bound to γEC by glutathione synthetase (GS) producing glutathione (GSH). Finally, trypanothione synthetase (TryS) synthesizes T(SH)2 by binding two GSH molecules to a Spd molecule. Trypanothione reductase (TyrR) catalyzes oxidized T(SH)2 reduction to TS2.
5. The Impact to Develop Inhibitors against Enzymes Involved in Spermidine Biosynthesis and Metabolism Treating Human African Trypanomiasis (HAT)
Currently, the only polyamine biosynthetic inhibitor that has been registered for HAT treatment is the irreversible ODC inhibitor DFMO
[25]. It is mostly administered in combination with other drugs (nifurtimox, suramin, melarsoprol) depending on the type of Trypanosomes (
T. gambiense or
T. rhodesiense) and the disease state of infection (presence or absence of central nervous symptoms). Generally, DFMO is used intravenously for late-stage
T. gambiense infection. However, the drug regimen is complicated and hampers patients’ compliance due to the high dosing course of 200 mg/kg for 7 days. DFMO also targets ODC from humans. However, selective toxicity to the enzyme from the parasite is achieved due to differences in intracellular turnover rates
[26]. Hence, pentamidine is still recommended for the first stage of
T. gambiense infection. Drug development for polyamine biosynthetic inhibitors in Trypanosomes is hampered by the fact that apart from the core polyamine pathway, a salvage pathway exists.
This entry is adapted from the peer-reviewed paper 10.3390/biom13050803