You're using an outdated browser. Please upgrade to a modern browser for the best experience.
Please note this is a comparison between Version 2 by Catherine Yang and Version 1 by José S. Câmara.
Prostate cancer (PCa) is the most frequently occurring type of malignant tumor and a leading cause of oncological death in men. PCa is very heterogeneous in terms of grade, phenotypes, and genetics, displaying complex features. Screening for PCa is based on the PSA biomarker values in blood serum (>4.0 ng/mL) and DRE. After suspicion, a magnetic resonance imaging (MRI) scan is usually performed, which indicates whether a prostate biopsy should be performed, considering the prostate imaging–reporting and data system (PI-RADS) value (PI-RADS > 3).
- prostate cancer
- incidence
- mortality
- risk factors
1. Prostate-Specific Membrane Antigen: A Theranostic Approach
Imaging methods are used to define the stage of PCa and so guide its management. However, PCa’s more aggressive forms can manifest rapid growth with progression to adjacent organs and spread to lymph nodes and bones [2,113[1][2][3],114], and CT, bone scan, and MRI have limited performance abilities in the detection of lymph node metastasis [92][4]. Patients with castration-resistant PCa (CRPCa) have a 90 to 95% probability of developing bone metastases, which leads to severe morbidity, including bone pain, pathological fractures, spinal cord compression, and hematological consequences of bone marrow infiltration [115,116,117][5][6][7]. Due to the importance of bone metastases in the overall disease progression, bone-targeted therapy constitutes an essential part of the treatment of CRPCa [118][8]. A possible therapy may be based on the use of radiopharmaceuticals systemically administered to slow or reverse the bone metastatic progression [117][7].
Current research is focused on the molecular targeting of prostate-specific membrane antigen (PSMA) as a theragnostic approach, to diagnose, monitor, and treat PCa [92][4]. PMSA is a transmembrane enzymatic protein found on most PCa cells, and its overexpression correlates to adverse factors, such as androgen independence, metastasis, and progression, making PSMA an antigenic marker for PCa progression [92,93,117,118][4][7][8][9]. Hence, PMSA can be used for diagnostic and therapeutic purposes, and several clinical trials have been investigating its effectiveness as a diagnostic tool and for direct radioligand therapy (Table 21) [92][4].
Table 21.
Emerging diagnostic methods for prostate cancer detection and management (articles from the last 5 years).
| Method | Evidence/Aim | Reference |
|---|---|---|
| PSMA radioligand targeted therapy and molecular imaging | Evidence: Molecular imaging techniques detect PCa lesions that are occult on anatomic imaging. PSMA radioligand therapy shows promising response rates with low toxicity in extensively pre-treated patients with PCa. Aim: Theragnostic applications—diagnosis, management, and treatment of metastatic PCa. |
[92,93,94,95,96,97,98,99,100][4][9][10][11][12][13][14][15][16] |
| EVs | Evidence: EVs can mediate PCa progression and metastasis. EVs have great potential to be used as liquid biopsy biomarkers in the diagnosis of PCa. EVs can be used in risk stratification and to predict the response to hormonal, chemo-, immune- and targeted therapy. Aim: Diagnosis and treatment. Can be used to personalize and guide treatment decisions. |
[76,87,89,101,102,103,104,105][17][18][19][20][21][22][23][24] |
| lncRNAs (PCA3, MALAT1, SChLAP1, BDNF-AS, FALEC) |
Evidence: lncRNAs provide new insights into cancer signaling networks, along with novel strategies and methods for PCa diagnosis and treatment. lncRNAs analysis has the potential to improve the specificity and sensitivity of existing biomarkers. Aim: Novel biomarkers (predictive, diagnostic, prognostic) and therapeutic targets. |
[106,107,108,109,110,111,112][25][26][27][28][29][30][31] |
Legend: EVs: extracellular vesicles; lncRNAs: long non-coding RNAs; PSMA: molecular targeting of prostate-specific membrane antigen.
1.1. Molecular Imaging
PSMA scans can detect metastatic lesions that are missed by conventional imaging techniques [92][4], so small molecules, antibodies, and antibody fragments that target PSMA have been created, radiolabeled, and used for molecular imaging [98][14].
PET is emerging as a highly sensitive molecular imaging technique in the detection and localization of primary PCa. PET uses a positron emitter to label key molecules that are intravenously injected, and their distribution and uptake images provide insights into metabolic changes associated with cancer [119][32]. This technique has been reported as a valuable tool in the diagnosis of PCa patients with negative MRI and systematic biopsies [98][14]. Recently, ligands of PSMA were introduced in PET to diagnose and manage PCa (reviewed by Mena et al., 2020 [99][15]). This approach can improve PCa detection by identifying lesions that are not visible on MRI, providing better estimates of tumor volume [98][14]. PSMA-PET can be used in the diagnosis, staging, and management of PCa patients [99][15]. PSMA-PET has an important role in the initial staging of PCa, superior diagnostic performance to anatomical imaging, and enhanced sensitivity to detect node metastasis (reaching 99% [119][32]), outperforming other molecule imaging techniques, including PET-CT [98,99][14][15]. Furthermore, PSMA-PET can be combined with anatomical CT (PET/CT) and MRI (PET/MRI) images for the detection of bone metastases [99,100][15][16] (Table 21). PSMA-PET/MRI consistently outperforms multiparametric MRI (mpMRI) in the detection or localization of PCa in intermediate- or high-risk PCa patients (reviewed by Moradi et al., 2021 [98][14] and Mena et al., 2020 [99][15]). PSMA-PET/CT has greater sensitivity in the detection of bone metastasis when compared to whole-body bone scans [100][16], and has shown the most utility in biochemical recurrence [119][32]. PSMA-PET/CT was first coupled with gallium-68 (68Ga) and is considered the most sensitive and specific method for staging high-risk PCa and imaging recurrent PCa [92,98][4][14]. Moreover, 68Ga-labeled ligands have shown higher sensitivity and specificity in the diagnosis of primary and recurrent PCa [100][16]. In a retrospective analysis, Maurer et al. [120][33] investigated the diagnostic efficacy of 68Ga-PSMA-PET for lymph node staging in patients with PCa and compared it to CT and MRI imaging. In their analyses, 68Ga-PSMA-11 showed sensitivity, specificity, and accuracy levels of 65.9%, 98.9%, and 88.5%, respectively, in the detection of nodal metastases, compared with the values of 43.9%, 85.4%, and 72.3% achieved by morphological imaging [120][33]. In another study, Thomas et al. [100][16] investigated the difference between technetium-99m (99mTc)-methyl diphosphate (MDP) bone scans and 68Ga-PSMA-PET/CT for the detection of bone metastases in PCa. The authors compared the number of identified lesions and found that the PSMA-PET/CT method detected twice the number of lesions, especially in the thorax and pelvis. Their results suggest that when patients go through 68Ga-PSMA-PET/CT, the bone scan is not mandatory [100][16].
1.2. Radioligand Targeted Therapy
Recent studies suggest that newer molecular theragnostic approaches, based on PSMA radioligands, have the potential to provide even more effective and personalized treatment options for diagnostic, prognostic, and therapeutic applications in patients with CRPCa, with fewer toxicities and adverse effects [92,93,94][4][9][10]. This approach has been developed to select patients, and delivers irradiation to all tumor sites, including osseous, nodal, and visceral metastases [92][4]. PSMA radioligand therapy uses small-molecule inhibitors of PSMA, usually labeled with beta and alpha-emitting radionuclides that emit cytotoxic radioactive decay [92,93][4][9]. Alpha and beta radionuclides differ in energy, tissue range, linear energy transfer, and the number of DNA hits needed for cell destruction [117][7]. These radiopharmaceuticals deliver targeted irradiation to the active bone turnover sites, where metastatic infiltration and destruction are happening. This approach can simultaneously treat multiple sites of disease, ease administration, and be integrated or combined with other treatments. Alpha-emitters include actinium-225 (225Ac), thorium-227 (227Th), radium-223 (223Ra), and astatine-211 (211At). Recently, 223Ra was approved to treat bone metastases from PCa. This authorization follows the symptomatic relief and significant improvement in the overall survival of CRPCa with predominant bone metastases that 223Ra was shown to elicit [121][34]. Beta-emitting radiopharmaceuticals, including lutetium-117 (177Lu), strontium-89 (89Sr), samarium-153 (153Sm), and rhenium-186 (186Re), have been used for bone palliation. 177Lu is the most used beta-emitter, due to its favorable safety profile, short range of emissions, and relatively long half-life, allowing the delivery of a high degree of radiation to specific lesions [92][4]. For instance, [177Lu] Lu-PSMA-617 shows a favorable safety profile due to reduced kidney uptake, and has demonstrated promising results in prospective trials with high response rates, low toxic effects, and the reduction of pain in men with metastatic CRPCa who progressed after standard treatments [95,96,97][11][12][13]. In general, radioligand therapy shows promising response rates with low toxicity in extensively pretreated patients with PCa [92][4]. While most of these studies remain experimental and the effects of this therapy on overall survival and safety are yet to be determined, their clinical observations are very promising [95,118,122,123,124][8][11][35][36][37].
PSMA-targeted imaging and therapy have proven to be excellent diagnostic and therapeutic options for metastatic PCa, but further studies are still required to determine the effect of this approach on overall survival and safety. Moreover, current research is still ongoing regarding the exact role of PSMA in various stages of PCa care [92][4].
2. Tumor Biomarkers
In recent years, new potential biomarkers for PCa screening and management have been developed through advances in molecular medicine, particularly OMICs genomics, proteomics, transcriptomics, and lipidomics. In addition to molecular biomarkers for urine, serum, and tissue samples, extracellular vesicles (EVs), circulating tumor cells (CTCs) and DNA (ctDNA), and cell-free DNA (cfDNA), common liquid biopsy biomarkers [125][38] and long noncoding ribonucleic acids (lncRNAs) have emerged as promising PCa biomarkers.
2.1. Molecular Biomarkers
Based on the combination of imaging techniques with other methodologies such as gene or protein profiling, several molecular biomarkers have been developed for urine, serum, and tissue samples to improve cancer detection, pre-biopsy decision-making, cancer risk assessment, and the therapeutic management of PCa [126][39]. Additionally, risk calculators (RCs) are used in combination with these tests to help identify each individual’s specific cancer risk, hence reducing the number of unnecessary biopsies. The guidelines on PCa treatment are therefore recommending the use of these tests in addition to the current PCa screening methods [77][40]. These biomarkers include several derivatives of PSA, such as the Prostate Health Index (PHI), approved by the US Food and Drug Administration (FDA), which combines total PSA, free PSA, and [−2] proPSA, and the Four-Kallikrein (4KScore) blood tests, which consist of kallikrein-related peptidase 2 (hK2), intact PSA, free PSA, and total PSA [104][23]. Transcriptomic methodologies also contributed to the discovery of biomarkers, and Progensa Prostate Cancer Antigen 3 (PCA3) is the first and only urine test approved by the FDA, which detects the PCa gene 3 transcript levels. The MyProstateScore (MPS) assay requires the collection of urine post-DRE and is based on combinations of multiple gene analyses, including total serum PSA, the PCA3 assay, and the expression of the TMPRSS2: ERG fusion gene [127,128][41][42]. These biomarkers can be used in liquid biopsies and involve a combination of clinical information, including age, family history, DRE result, PSA levels, and prostate biopsy history, with genetic and epigenetic changes. Nevertheless, the technologies associated with these approaches are expensive and unavailable in many medical facilities. Other factors such as tumor heterogeneity, tumor–host interplay, complexity, multiplicity, and redundancy of tumor–cell signaling networks must be overcome to develop effective biomarkers [81][43].
2.2. Long Non-Coding RNAs
LncRNAs are RNA transcripts that are longer than 200 nucleotides and do not encode proteins. LncRNAs have been found to exhibit abnormal expression in various types of cancer, including PCa. Most lncRNAs linked to PCa are overexpressed in tumor tissues and cancer cells, contributing to tumor proliferation, invasion, and metastasis. In turn, only a small number of lncRNAs are downregulated and may function as tumor suppressors in addition to their roles as transcriptional regulators and oncogenes [106][25]. All these unique features make lncRNAs promising prognostic biomarkers and therapeutic targets for the diagnosis, screening, prognosis, and progression of PCa [106][25] (Table 21). Recent research has demonstrated that lncRNAs such as PCA3, GAS5, and HOTAIR are associated with the development and progression of PCa [106][25]. Given its higher specificity and sensitivity than the PSA blood test, PCA3 is one of the most well-studied lncRNAs. Additionally, its combination with PSA testing or other biomarkers will significantly improve the sensitivity, specificity, and accuracy of PCa screening and diagnosis. For instance, the use of PCA3 in conjunction with TMPRSS2-ERG tests can reduce the number of unnecessary biopsies and increase diagnostic accuracy [106][25]. Another putative PCa diagnostic marker is MALAT1, whose increased expression has been linked to high PSA levels and Gleason scores, as well as with tumor stage and CRPCa [106][25]. Single-nucleotide polymorphisms of MALAT1 were investigated by Hu et al. [109][28], who found that rs619586 and rs1194338 were significantly associated with PCa’s susceptibility to both advanced Gleason grade and nodal metastasis. A noninvasive post-DRE urine assay based on the combination of the lncRNAs PCA3 and MALAT1 for the early diagnosis of PCa and high-grade tumors was developed and validated by Li and collaborators [110][29]. However, according to some researchers, the PCA3 test is affected by intra-individual variability, being unable to differentiate between high-grade and low-grade tumors. Hence, more data are necessary to determine PCA3’s application in PCa diagnosis [106][25]. The lncRNAs TMPO-AS1 and FALEC have shown their potential utility as biomarkers for PCa diagnosis and progression [106,112][25][31]. Zhao et al. [108][27] examined the biological role of FALEC in PCa cell lines as well as its expression profile, and paired histologically normal tissues. In 85 patients, clinical PCa tissues showed significantly higher FALEC expressions when compared to adjacent normal tissues. Moreover, in vitro cell proliferation, migration, and invasion could be inhibited by the downregulation of FALEC. According to these findings, FALEC may be a useful diagnostic and therapeutic target in PCa patients [108][27]. Li et al. [107][26] investigated the expression, prognostic value, and functional role of lncRNA BDNF-AS in PCa. The authors also correlated the expression of BDNF-AS with the clinicopathological factors of patients. The results of this study demonstrate the potential use of BDNF-AS as a prognostic biomarker for PCa patients with poor prognoses and shorter overall survival, as it was downregulated in these cases. Furthermore, lncRNAs can be used to predict the recurrence of biochemical events. SChLAP1 was highly expressed in PCa tissue, which was substantially correlated with biochemical recurrence, clinical progression, and PCa-specific mortality [111][30]. Additionally, SChLAP1 can be easily detected in urine, an important feature for the development of an SChLAP1 assay for guided therapy (as reviewed by Xu et al. [106][25]). Given the roles of lncRNAs in PCa, it will be important to create specific drugs that interfere with malignant signaling networks in which lncRNAs are engaged, particularly in PCa cells. However, it is still unclear how exactly lncRNAs work at the molecular level, it being essential to further investigate the role of lncRNAs in prostate carcinogenesis [106][25].
2.3. Liquid Biopsy Biomarkers
Liquid biopsy has emerged as a complement to invasive tissue biopsy to guide cancer diagnosis and treatment [76][17]. Liquid biopsies rely on the detection of specific biomarkers in readily accessible body fluids, such as blood, serum, or urine [89][19]. The common liquid biopsy biomarkers are EVs, CTCs, ctDNA, and cfDNA, which provide specific information based on their intrinsic characteristics. CTCs are cancer cells from primary and metastatic tumors that are released into the vasculature and circulate through the body to form metastatic niches in other tissues, being detectable in cancer patients only [125][38]. Similarly, ctDNA is a tumor-derived short, fragmented DNA found in the bloodstream, which reflects cancer-related genetic changes. cfDNA or RNA (cfRNA) are cell-free circulating small nucleic acid fragments that are released after the lysis of apoptotic or necrotic cells. cfDNA is detectable in blood and urine samples from patients with cancer, and their analyses improve the evaluation of mutations, polymorphism, methylation, and loss of DNA integrity [76,89,129][17][19][44]. Numerous studies have shown the relevance of liquid biopsies in PCa screening. cfDNA and EVs seem to have a better application in the diagnosis and prognosis of PCa than CTCs [76,87,89,101][17][18][19][20] (Table 21). This occurs because early-stage or localized PCa patients have very few CTCs and their use is more effective in the later stages of this cancer [89][19]. The only FDA-approved liquid biopsy test for PCa, CellSearch, is based on the detection of CTCs, and there is no evidence of the wide clinical implementation of this technology in medical practice. EVs are nano-sized, double-lipid membrane vesicles, such as exosomes and microvesicles, that are secreted from cells and shed into biofluids, including blood and urine [104][23]. EVs are involved in intercellular communication and immune function, through proteins, lipids, mRNA, microRNAs (miRNAs), and DNA, and have been correlated to the presence of cancer for diagnostic purposes (Table 21) [76,101,130,131][17][20][45][46]. Cells exchange proteins, nucleic acids, sugars, and lipids through EVs to induce changes in the recipient cells, which makes EVs potential carriers of cancer biomarkers from tumor cells to other tumor or non-tumor cells [89][19]. EVs can also be used as a vehicle for drugs or nucleic acids with antineoplastic effects [87,102][18][21]. The EVs approach may improve the sensitivity of PCa biomarkers, given the protective role of the EVs’ lipid layer over biomolecules, meaning that the concentration of PCa biomarkers will be higher in EVs [89][19]. Urine is the most used body fluid for the detection of biomarkers in EVs from liquid biopsies of PCa. Moreover, exosomal miRNAs are emerging as promising prognostic biomarkers for metastatic CRPCa patients [89][19]. The concentration of RNA-based biomarkers, particularly miRNA, is higher in EVs than in CTCs from urine samples. Nevertheless, the application of miRNA as a diagnostic marker has been limited due to a lack of specificity, and in turn, many studies have emerged to investigate EV-mRNA as a diagnostic and prognostic biomarker for PCa management [76][17]. McKiernan et al. [104][23] developed an exosome-derived gene expression signature from normalized PCA3 and ERG RNA from urine predictive of initial biopsy results. Exosomes in post-DRE urine of PCa patients contain both PCA3 and TMPRSS2: ERG mRNA. In their study, the authors were able to develop a molecular signature predictive of PCa combined with serum PSA in a diagnostic test, which was able to discriminate between benign disease and high- and low-grade tumors, reducing the total number of unnecessary biopsies [104][23]. Ji et al. [105][24] developed a strategy for exosomal mRNA detection based on features of mRNA of circulating exosomes and identified a PCa exosomal mRNA signature for PCa screening and diagnosis. With this strategy, the authors were able to distinguish PCa patients from healthy controls [105][24]. Despite the beneficial properties of EVs for the diagnosis of PCa, their clinical application still presents a few challenging issues [76][17]. EVs are released from all cells in the body, which makes it difficult to determine which EVs are tumor-derived, meaning that new technologies for the specific detection and isolation of tumor-derived EVs need to be developed [76][17]. Recent EVs isolation technologies have been developed to improve isolation performance, yield, purity, usability, hands-on procedures, and processing time [76][17]. However, EVs isolation is still difficult, especially in EVs from blood plasma, due to the purity and efficiency achieved by laboratory procedures. Moreover, there is no wide clinical application of liquid biopsies of PCa with EVs [89][19], and automated analysis platforms are yet to be developed for large-scale clinical studies [76][17]. Overall, the use of CTCs and EVs as biomarkers of PCa in liquid biopsies is being hindered by some issues, such as the inexistence of specific guidelines for the biomarker’s isolation and detection. Additionally, the validation and standardization of the microfluidic devices used in liquid biopsies has not been achieved yet [129][44].
3. Active Surveillance and Risk-Stratification Algorithms
PCa is very heterogeneous in terms of grade, phenotypes, and genetics, displaying complex features [2][1]. This tumor often has indolent growth, which does not compromise the patient’s quality of life, but its diagnosis and subsequent treatments have a high impact on the physical and mental status of patients, significantly affecting their quality of life [81][43]. The main goal of early detection is to identify PCa in a phase whereat it needs less aggressive treatments with fewer side effects and has a higher chance of cure, even in the cases of locally advanced and metastatic PCa. Many early diagnoses can be safely managed by active surveillance, preventing overtreatment, thereby improving or maintaining the patient’s quality of life and avoiding adverse outcomes [132][47].
Active surveillance consists of the serial monitoring of disease progression, through PSA tests, DRE, and biopsies, to track cancer growth. This has become the preferred approach for men with low-grade PCa [2[1][48],133], as men can avoid immediate treatment and prospective side effects [2][1]. When discussing therapy choices and in the selection criteria for active surveillance programs [134][49], external factors, such as obesity, BMI, and the hormonal profile (e.g., testosterone levels), should be considered by the clinical practice, since all these factors influence the PSA levels [135,136][50][51]. Recent studies suggest that the conjugation of PSA screening with other methodologies, such as risk RCs, biomarkers, and imaging techniques such as MRI, can attenuate overdiagnosis and underdetection issues [137][52]. Van Poppel et al. [137][52] proposed a risk-stratified algorithm, combining MRI, RC, and PSA tests, that improves the efficiency of “PSA-only” screening and reduces unnecessary biopsies and overdiagnosis. The combination of these tools improves the individual balance between the harms and benefits of early detection in well-informed men who are at risk of having PCa [137][52]. Based on the initial PSA test result and age, different time intervals for repeated PSA testing are proposed, reflecting the likelihood of a future diagnosis of clinically significant cancer. This strategy helps to avoid false-positive biopsies, as low-risk men can go through individualized PSA tests and, if necessary, repeated MRIs to track cancer growth. Then, RCs seem to be the most appropriate approach to assessing the risk of developing PCa after PSA testing. RCs are accessible to every clinician, easy to use, inexpensive, and non-invasive. Moreover, MRI results can be integrated into an RC that includes PSA density as a continuous variable, to determine the need for a prostate biopsy in men with intermediate- and high-risk [137,138][52][53]. PSA density has been described to improve the specificity of the PSA test [138,139][53][54]. It is defined as the level of serum PSA divided by the prostate volume and presents a cut-off of 0.15 ng/mL2 [139][54]. PSA density can be used as a prognostic biomarker to determine which patients need to undergo definitive therapy from those who may be managed by active surveillance, as well as patients with a previously negative MRI who should proceed to a prostate biopsy [139][54]. This allows the more accurate evaluation of individual risk, which is essential for properly interpreting the MRI results. Consequently, only men who present a high risk of clinically significant PCa, according to an RC, will be proposed for a systemic biopsy after MRI [137][52].
Evidence shows that performing an MRI before a biopsy allows one-third of men to avoid an immediate biopsy and reduces overdiagnosis, with 40% fewer clinically unimportant cancers and approximately 15% more clinically significant cancers detected [137,140][52][55]. However, the implementation of MRI in the risk assessment of PCa is not yet fully realized in the whole of Europe [137][52], which in turn reflects the geographical differences in the incidence rates between European countries. To further reduce unnecessary biopsy procedures, the decision process of a biopsy in men with a PI-RADS of 3 should be carefully examined. The PI-RADS classification is based on a scale of values from 1 to 5, and determines the likelihood of clinically significant PCa. While PI-RADS values of 4 and 5 indicate that a biopsy is required, it is challenging to establish whether a biopsy should be performed or not in patients with a score of 3 [141][56]. Additionally, the PI-RADS score does not measure PCa aggressiveness, meaning that a biopsy is still needed. Research has found that excluding men with PI-RADS 1–2 or PI-RADS 3 lesions based on a low PSA density only increases the likelihood that clinically significant tumors will be undiagnosed due to nonvisual PCa or misinterpretation of the reader [137][52]. The European Association of Urology (EAU) guidelines strongly recommend performing an mpMRI before a biopsy to modify the management approach accordingly. This imaging approach presents preferable detection rates and reduces the number of biopsy procedures, particularly when MRI-negative men are excluded from prostate biopsy, due to its capacity to differentiate between significant and insignificant tumors [132][47]. Furthermore, the PI-RADS guidelines have recommended systematized mpMRI acquisition and the global standardization of reporting. Nevertheless, there is a lack of consensus on detailed aspects of mpMRI acquisition protocols [141][56].
Artificial intelligence (AI) methods have been proposed for a wide range of applications in the PCa diagnostic pathway [137,141,142,143][52][56][57][58]. AI can be used to improve the initial evaluation of prostate mpMRI cases and the image quality, as well as the detection and differentiation of clinically significant from insignificant cancers on a voxel level, and the classification of entire lesions into PI-RADS categories (reviewed by Belue and Turkbey [142][57] and Sunoqrot et al. [143][58]). Studies on MRI AI have revealed the role of AI in improving the clinical management of localized PCa, the interpretation of MRI and the data processing for biopsies, by reducing inter-reader variation and supporting the radiological workflow [142][57]. Nevertheless, AI requires caution in its use, as the proficiency of this method is still below that of an expert [141][56]. Moreover, more prospective studies with multicenter designs are required to understand the impact of AI on improving radiologists’ performance and the clinical management of PCa [137,142][52][57].
4. Volatilomics
Emerging studies demonstrate that combining PSA screening with other methodologies, such as RCs, biomarkers, and imaging tests, e.g., MRI or fusion biopsies, might attenuate overdiagnosis and underdetection, eventually reducing the number of unnecessary biopsies [137][52]. Volatilomics, a subset of metabolomics, has recently emerged as a simple, effective, and non-invasive method with great potential for cancer screening. Volatilomics focuses on volatile organic metabolites (VOMs), which are low-molecular weight metabolites (<500 Da) with high volatility and a carbon-based chemical group [144][59]. VOMs are present in readily accessible biofluids, including saliva, urine, and exhaled breath, as they are produced by the metabolism of cells, reflecting their biological activity [145][60]. The progressive accumulation of genetic, epigenetic, and post-translational changes that support cancer growth can lead to changes in VOMs levels and, as a result, affect an individual’s volatilomic profile (Figure 51). Hence, VOMs are a rich source of data on health, since they can reflect the metabolic and biochemical alterations triggered by cancer progression. From this perspective, a volatilomic biosignature for diagnostic purposes can be defined using these changes [77,86][40][61].
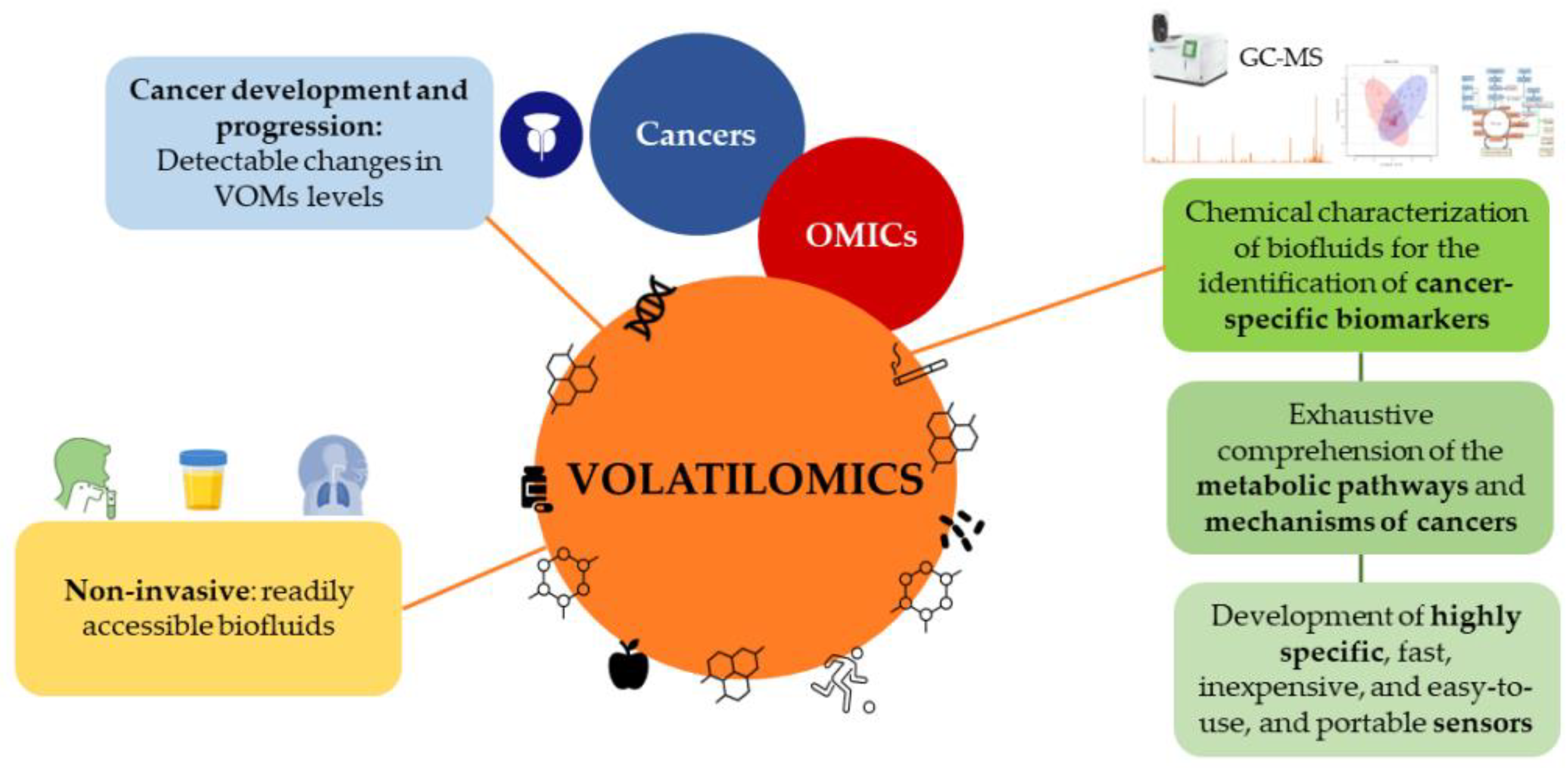
Figure 51. Cancer development and progression can lead to changes in the levels of volatile organic metabolites, which can be used to define a volatilomic biosignature for diagnostic purposes.
Even though the volatilomics approach is relatively recent in PCa compared to other cancers [77[40][62][63][64],88,146,147], empirical data have confirmed its potential use in cancer screening, the monitoring of disease progression and effectiveness of treatment, as well as for the discrimination between different cancer types [86,148,149,150][61][65][66][67]. Different approaches involving volatilomic studies have been proposed to establish connections between cancer and the body’s VOMs signature using highly sensitive analytical techniques. In these studies, biofluids are chemically characterized to identify cancer-specific biomarkers using mass spectrometry-based techniques combined with multivariate statistical analysis. Another approach includes the identification of cancer-characteristic odor fingerprints through electronic noses (e-noses) [151][68]. However, since several VOMs have been suggested as PCa biomarkers and contradictory results on the same metabolites have emerged from different reports, it is difficult to establish reliable biomarkers, and no exhaustive studies have yet been published [151,152][68][69]. Additionally, a few restrictions hinder the implementation of these approaches in real-time diagnostic applications, and consequently, in clinical practice (reviewed by Berenguer et al. [144][59]). For instance, the ability to compare the outcomes of various studies between different laboratories is hampered by variations in sample preparation, analytical procedures, and statistical platforms [88][62]. Hence, methods must be standardized from sample collection to data processing, as well as assess the impact of confounding factors, such as epigenetics, diet, medication, genetics, and environmental exposure. Epigenetic factors play an important role in determining the clinical phenotypes of PCa. Therefore, due to genetic, environmental, and toxicological factors, as well as the different dietary habits around the world and their influence on the development of cancer, the volatilomic biosignatures and potential biomarkers will differ according to the region of the world [77,88,144,146,147][40][59][62][63][64].
Despite these limitations, volatilomics offers a wealth of informational potential that will allow a thorough understanding of the metabolic pathways, and a clarification of the mechanisms of cancers and how they impact the generation of VOMs [153][70]. Further analysis of the VOMs’ origin and a more accurate assessment of the impact of confounding factors on the volatilomic profile will be possible as a result of these findings [147][64]. Additionally, the definition of cancer biomarkers will be made possible through the detection and quantification of specific metabolites due to the standardization of procedures and the creation of highly focused sensors. These findings will foster the development of highly specific, fast, inexpensive, easy-to-use, and portable sensors that can be implemented in clinical practice [145[60][71],154], demonstrating the importance of the volatilomics approach [151,155][68][72]. Hopefully, the progress in volatilomics studies will unveil biomarkers suitable for the diagnosis of PCa, to be used as a supplement to the current approaches for the classification and screening of cancer [129][44], with possible applications in the active surveillance of patients and individualized care [81,144][43][59].
References
- Rawla, P. Epidemiology of Prostate Cancer. World J. Oncol. 2019, 10, 63–89.
- Beltran, H.; Demichelis, F. Intrapatient heterogeneity in prostate cancer. Nat. Rev. Urol. 2015, 12, 430–431.
- Dudka, I.; Thysell, E.; Lundquist, K.; Antti, H.; Iglesias-Gato, D.; Flores-Morales, A.; Bergh, A.; Wikström, P.; Gröbner, G. Comprehensive metabolomics analysis of prostate cancer tissue in relation to tumor aggressiveness and TMPRSS2-ERG fusion status. BMC Cancer 2020, 20, 437.
- Parsi, M.; Desai, M.H.; Desai, D.; Singhal, S.; Khandwala, P.M.; Potdar, R.R. PSMA: A game changer in the diagnosis and treatment of advanced prostate cancer. Med. Oncol. 2021, 38, 89.
- Coleman, R.E.; Lipton, A.; Roodman, G.D.; Guise, T.A.; Boyce, B.F.; Brufsky, A.M.; Clézardin, P.; Croucher, P.I.; Gralow, J.R.; Hadji, P.; et al. Metastasis and bone loss: Advancing treatment and prevention. Cancer Treat. Rev. 2010, 36, 615–620.
- Parker, C.; Nilsson, S.; Heinrich, D.; Helle, S.I.; O’Sullivan, J.M.; Fosså, S.D.; Chodacki, A.; Wiechno, P.; Logue, J.; Seke, M.; et al. Alpha Emitter Radium-223 and Survival in Metastatic Prostate Cancer. N. Engl. J. Med. 2013, 369, 213–223.
- Maffioli, L.; Florimonte, L.; Costa, D.; Castanheira Correira, J.; Grana, C.; Luster, M.; Bodei, L.; Chinol, M. New radiopharmaceutical agents for the treatment of castration-resistant prostate cancer. Q. J. Nucl. Med. Mol. Imaging 2015, 59, 420–438.
- Du, Y.; Dizdarevic, S. Molecular radiotheragnostics in prostate cancer. Clin. Med. J. R. Coll. Physicians Lond. 2017, 17, 458–461.
- Uijen, M.J.M.; Derks, Y.H.W.; Merkx, R.I.J.; Schilham, M.G.M.; Roosen, J.; Privé, B.M.; van Lith, S.A.M.; van Herpen, C.M.L.; Gotthardt, M.; Heskamp, S.; et al. PSMA radioligand therapy for solid tumors other than prostate cancer: Background, opportunities, challenges, and first clinical reports. Eur. J. Nucl. Med. Mol. Imaging 2021, 48, 4350–4368.
- Seifert, R.; Alberts, I.L.; Afshar-Oromieh, A.; Rahbar, K. Prostate Cancer Theranostics: PSMA Targeted Therapy. PET Clin. 2021, 16, 391–396.
- Hofman, M.S.; Violet, J.; Hicks, R.J.; Ferdinandus, J.; Thang, S.P.; Akhurst, T.; Iravani, A.; Kong, G.; Ravi Kumar, A.; Murphy, D.G.; et al. -PSMA-617 radionuclide treatment in patients with metastatic castration-resistant prostate cancer (LuPSMA trial): A single-centre, single-arm, phase 2 study. Lancet Oncol. 2018, 19, 825–833.
- Hofman, M.; Violet, J.A.; Hicks, R.J.; Ferdinandus, J.; Thang, S.P.; Iravani, A.; Kong, G.; Ravi Kumar, A.; Akhurst, T.J.; Mooi, J.; et al. Results of a 50 patient single-center phase II prospective trial of Lutetium-177 PSMA-617 theranostics in metastatic castrate-resistant prostate cancer. J. Clin. Oncol. 2019, 37, 228.
- Calais, J.; Fendler, W.P.; Eiber, M.; Lassmann, M.; Dahlbom, M.; Esfandiari, R.; Gartmann, J.; Nguyen, K.; Thin, P.; Lok, V.; et al. RESIST-PC phase 2 trial: 177Lu-PSMA-617 radionuclide therapy for metastatic castrate-resistant prostate cancer. J. Clin. Oncol. 2019, 37, 5028.
- Moradi, F.; Farolfi, A.; Fanti, S.; Iagaru, A. Prostate cancer: Molecular imaging and MRI. Eur. J. Radiol. 2021, 143, 109893.
- Mena, E.; Black, P.C.; Rais-Bahrami, S.; Gorin, M.; Allaf, M.; Choyke, P. Novel PET imaging methods for prostate cancer. World J. Urol. 2021, 39, 687–699.
- Thomas, L.; Balmus, C.; Ahmadzadehfar, H.; Essler, M.; Strunk, H.; Bundschuh, R.A. Assessment of Bone Metastases in Patients with Prostate Cancer-A Comparison between (99m)Tc-Bone-Scintigraphy and Ga-PSMA PET/CT. Pharmaceuticals (Basel) 2017, 10, 68.
- Kim, C.-J.; Dong, L.; Amend, S.R.; Cho, Y.-K.; Pienta, K.J. The role of liquid biopsies in prostate cancer management. Lab A Chip 2021, 21, 3263–3288.
- Gaglani, S.; Gonzalez-Kozlova, E.; Lundon, D.J.; Tewari, A.K.; Dogra, N.; Kyprianou, N. Exosomes as A Next-Generation Diagnostic and Therapeutic Tool in Prostate Cancer. Int. J. Mol. Sci. 2021, 22, 10131.
- Campos-Fernández, E.; Barcelos, L.S.; de Souza, A.G.; Goulart, L.R.; Alonso-Goulart, V. Research landscape of liquid biopsies in prostate cancer. Am. J. Cancer Res. 2019, 9, 1309–1328.
- Oey, O.; Ghaffari, M.; Li, J.J.; Hosseini-Beheshti, E. Application of extracellular vesicles in the diagnosis and treatment of prostate cancer: Implications for clinical practice. Crit. Rev. Oncol. Hematol. 2021, 167, 103495.
- Ludwig, M.; Rajvansh, R.; Drake, J.M. Emerging Role of Extracellular Vesicles in Prostate Cancer. Endocrinology 2021, 162, bqab139.
- Lorenc, T.; Klimczyk, K.; Michalczewska, I.; Słomka, M.; Kubiak-Tomaszewska, G.; Olejarz, W. Exosomes in Prostate Cancer Diagnosis, Prognosis and Therapy. Int. J. Mol. Sci. 2020, 21, 2118.
- McKiernan, J.; Donovan, M.J.; O’Neill, V.; Bentink, S.; Noerholm, M.; Belzer, S.; Skog, J.; Kattan, M.W.; Partin, A.; Andriole, G.; et al. A Novel Urine Exosome Gene Expression Assay to Predict High-grade Prostate Cancer at Initial Biopsy. JAMA Oncol. 2016, 2, 882–889.
- Ji, J.; Chen, R.; Zhao, L.; Xu, Y.; Cao, Z.; Xu, H.; Chen, X.; Shi, X.; Zhu, Y.; Lyu, J.; et al. Circulating exosomal mRNA profiling identifies novel signatures for the detection of prostate cancer. Mol. Cancer 2021, 20, 58.
- Xu, Y.H.; Deng, J.L.; Wang, G.; Zhu, Y.S. Long non-coding RNAs in prostate cancer: Functional roles and clinical implications. Cancer Lett. 2019, 464, 37–55.
- Li, W.; Dou, Z.; We, S.; Zhu, Z.; Pan, D.; Jia, Z.; Liu, H.; Wang, X.; Yu, G. Long noncoding RNA BDNF-AS is associated with clinical outcomes and has functional role in human prostate cancer. Biomed. Pharmacother. 2018, 102, 1105–1110.
- Zhao, R.; Sun, F.; Bei, X.; Wang, X.; Zhu, Y.; Jiang, C.; Zhao, F.; Han, B.; Xia, S. Upregulation of the long non-coding RNA FALEC promotes proliferation and migration of prostate cancer cell lines and predicts prognosis of PCa patients. Prostate 2017, 77, 1107–1117.
- Hu, J.C.; Wang, S.S.; Chou, Y.E.; Chiu, K.Y.; Li, J.R.; Chen, C.S.; Hung, S.C.; Yang, C.K.; Ou, Y.C.; Cheng, C.L.; et al. Associations between LncRNA MALAT1 Polymorphisms and Lymph Node Metastasis in Prostate Cancer. Diagnostics (Basel) 2021, 11, 1692.
- Li, Y.; Ji, J.; Lyu, J.; Jin, X.; He, X.; Mo, S.; Xu, H.; He, J.; Cao, Z.; Chen, X.; et al. A Novel Urine Exosomal lncRNA Assay to Improve the Detection of Prostate Cancer at Initial Biopsy: A Retrospective Multicenter Diagnostic Feasibility Study. Cancers (Basel) 2021, 13, 4075.
- Kidd, S.G.; Carm, K.T.; Bogaard, M.; Olsen, L.G.; Bakken, A.C.; Løvf, M.; Lothe, R.A.; Axcrona, K.; Axcrona, U.; Skotheim, R.I. High expression of SCHLAP1 in primary prostate cancer is an independent predictor of biochemical recurrence, despite substantial heterogeneity. Neoplasia 2021, 23, 634–641.
- Huang, W.; Su, X.; Yan, W.; Kong, Z.; Wang, D.; Huang, Y.; Zhai, Q.; Zhang, X.; Wu, H.; Li, Y.; et al. Overexpression of AR-regulated lncRNA TMPO-AS1 correlates with tumor progression and poor prognosis in prostate cancer. Prostate 2018, 78, 1248–1261.
- Retter, A.; Gong, F.; Syer, T.; Singh, S.; Adeleke, S.; Punwani, S. Emerging methods for prostate cancer imaging: Evaluating cancer structure and metabolic alterations more clearly. Mol. Oncol. 2021, 15, 2565–2579.
- Maurer, T.; Gschwend, J.E.; Rauscher, I.; Souvatzoglou, M.; Haller, B.; Weirich, G.; Wester, H.J.; Heck, M.; Kübler, H.; Beer, A.J.; et al. Diagnostic Efficacy of (68)Gallium-PSMA Positron Emission Tomography Compared to Conventional Imaging for Lymph Node Staging of 130 Consecutive Patients with Intermediate to High Risk Prostate Cancer. J. Urol. 2016, 195, 1436–1443.
- Cursano, M.C.; Iuliani, M.; Casadei, C.; Stellato, M.; Tonini, G.; Paganelli, G.; Santini, D.; De Giorgi, U. Combination radium-223 therapies in patients with bone metastases from castration-resistant prostate cancer: A review. Crit. Rev. Oncol. Hematol. 2020, 146, 102864.
- Afshar-Oromieh, A.; Babich, J.W.; Kratochwil, C.; Giesel, F.L.; Eisenhut, M.; Kopka, K.; Haberkorn, U. The Rise of PSMA Ligands for Diagnosis and Therapy of Prostate Cancer. J. Nucl. Med. 2016, 57, 79S–89S.
- Rahbar, K.; Ahmadzadehfar, H.; Kratochwil, C.; Haberkorn, U.; Schäfers, M.; Essler, M.; Baum, R.P.; Kulkarni, H.R.; Schmidt, M.; Drzezga, A.; et al. German Multicenter Study Investigating 177Lu-PSMA-617 Radioligand Therapy in Advanced Prostate Cancer Patients. J. Nucl. Med. 2017, 58, 85–90.
- Scarpa, L.; Buxbaum, S.; Kendler, D.; Fink, K.; Bektic, J.; Gruber, L.; Decristoforo, C.; Uprimny, C.; Lukas, P.; Horninger, W.; et al. The (68)Ga/(177)Lu theragnostic concept in PSMA targeting of castration-resistant prostate cancer: Correlation of SUV(max) values and absorbed dose estimates. Eur. J. Nucl. Med. Mol. Imaging 2017, 44, 788–800.
- Morrison, G.J.; Goldkorn, A. Development and Application of Liquid Biopsies in Metastatic Prostate Cancer. Curr. Oncol. Rep. 2018, 20, 35.
- Matuszczak, M.; Schalken, J.A.; Salagierski, M. Prostate Cancer Liquid Biopsy Biomarkers’ Clinical Utility in Diagnosis and Prognosis. Cancers 2021, 13, 3373.
- Lima, A.R.; Pinto, J.; Amaro, F.; Bastos, M.d.L.; Carvalho, M.; de Pinho, P.G. Advances and Perspectives in Prostate Cancer Biomarker Discovery in the Last 5 Years through Tissue and Urine Metabolomics. Metabolites 2021, 11, 181.
- Bae, J.; Yang, S.H.; Kim, A.; Kim, H.G. RNA-based biomarkers for the diagnosis, prognosis, and therapeutic response monitoring of prostate cancer. Urol. Oncol. 2022, 40, 105.e1–105.e10.
- Kan, Y.; Li, B.; Yang, D.; Liu, Y.; Liu, J.; Yang, C.; Mao, L. Emerging Roles of Long Non-coding RNAs as Novel Biomarkers in the Diagnosis and Prognosis of Prostate Cancer. Discov. Med. 2021, 32, 29–37.
- Salciccia, S.; Capriotti, A.L.; Lagana, A.; Fais, S.; Logozzi, M.; De Berardinis, E.; Busetto, G.M.; Di Pierro, G.B.; Ricciuti, G.P.; Del Giudice, F.; et al. Biomarkers in Prostate Cancer Diagnosis: From Current Knowledge to the Role of Metabolomics and Exosomes. Int. J. Mol. Sci. 2021, 22, 4367.
- Kretschmer, A.; Tilki, D. Biomarkers in prostate cancer—Current clinical utility and future perspectives. Crit. Rev. Oncol. Hematol. 2017, 120, 180–193.
- Hanjani, N.A.; Esmaelizad, N.; Zanganeh, S.; Gharavi, A.T.; Heidarizadeh, P.; Radfar, M.; Omidi, F.; MacLoughlin, R.; Doroudian, M. Emerging role of exosomes as biomarkers in cancer treatment and diagnosis. Crit. Rev. Oncol. Hematol. 2022, 169, 103565.
- Wang, J.; Ni, J.; Beretov, J.; Thompson, J.; Graham, P.; Li, Y. Exosomal microRNAs as liquid biopsy biomarkers in prostate cancer. Crit. Rev. Oncol. Hematol. 2020, 145, 102860.
- Van Poppel, H.; Roobol, M.J.; Chapple, C.R.; Catto, J.W.F.; N’Dow, J.; Sønksen, J.; Stenzl, A.; Wirth, M. Prostate-specific Antigen Testing as Part of a Risk-Adapted Early Detection Strategy for Prostate Cancer: European Association of Urology Position and Recommendations for 2021. Eur. Urol. 2021, 80, 703–711.
- Litwin, M.S.; Tan, H.J. The diagnosis and treatment of prostate cancer: A review. JAMA J. Am. Med. Assoc. 2017, 317, 2532–2542.
- Ossoliński, K.; Nizioł, J.; Arendowski, A.; Ossolińska, A.; Ossoliński, T.; Kucharz, J.; Wiechno, P.; Ruman, T. Mass spectrometry-based metabolomic profiling of prostate cancer—A pilot study. J. Cancer Metastasis Treat. 2019, 5, 1.
- de Cobelli, O.; Terracciano, D.; Tagliabue, E.; Raimondi, S.; Galasso, G.; Cioffi, A.; Cordima, G.; Musi, G.; Damiano, R.; Cantiello, F.; et al. Body mass index was associated with upstaging and upgrading in patients with low-risk prostate cancer who met the inclusion criteria for active surveillance. Urol. Oncol. 2015, 33, 201.e1–208.e8.
- Ferro, M.; Lucarelli, G.; Bruzzese, D.; Di Lorenzo, G.; Perdonà, S.; Autorino, R.; Cantiello, F.; La Rocca, R.; Busetto, G.M.; Cimmino, A.; et al. Low serum total testosterone level as a predictor of upstaging and upgrading in low-risk prostate cancer patients meeting the inclusion criteria for active surveillance. Oncotarget 2017, 8, 18424–18434.
- Van Poppel, H.; Hogenhout, R.; Albers, P.; van den Bergh, R.C.N.; Barentsz, J.O.; Roobol, M.J. A European Model for an Organised Risk-stratified Early Detection Programme for Prostate Cancer. Eur. Urol. Oncol. 2021, 4, 731–739.
- Yusim, I.; Krenawi, M.; Mazor, E.; Novack, V.; Mabjeesh, N.J. The use of prostate specific antigen density to predict clinically significant prostate cancer. Sci. Rep. 2020, 10, 20015.
- Omri, N.; Kamil, M.; Alexander, K.; Alexander, K.; Edmond, S.; Ariel, Z.; David, K.; Gilad, A.E.; Azik, H. Association between PSA density and pathologically significant prostate cancer: The impact of prostate volume. Prostate 2020, 80, 1444–1449.
- Drost, F.J.H.; Osses, D.F.; Nieboer, D.; Steyerberg, E.W.; Bangma, C.H.; Roobol, M.J.; Schoots, I.G. Prostate MRI, with or without MRI-targeted biopsy, and systematic biopsy for detecting prostate cancer. Cochrane Database Syst. Rev. 2019, 6, CD012663.
- Gravina, M.; Spirito, L.; Celentano, G.; Capece, M.; Creta, M.; Califano, G.; Collà Ruvolo, C.; Morra, S.; Imbriaco, M.; Di Bello, F.; et al. Machine Learning and Clinical-Radiological Characteristics for the Classification of Prostate Cancer in PI-RADS 3 Lesions. Diagnostics (Basel) 2022, 12, 1565.
- Belue, M.J.; Turkbey, B. Tasks for artificial intelligence in prostate MRI. Eur. Radiol. Exp. 2022, 6, 33.
- Sunoqrot, M.R.S.; Saha, A.; Hosseinzadeh, M.; Elschot, M.; Huisman, H. Artificial intelligence for prostate MRI: Open datasets, available applications, and grand challenges. Eur. Radiol. Exp. 2022, 6, 35.
- Berenguer, C.V.; Pereira, F.; Pereira, J.A.M.; Câmara, J.S. Volatilomics: An Emerging and Promising Avenue for the Detection of Potential Prostate Cancer Biomarkers. Cancers 2022, 14, 3982.
- Janfaza, S.; Khorsand, B.; Nikkhah, M.; Zahiri, J. Digging deeper into volatile organic compounds associated with cancer. Biol. Methods Protoc. 2019, 4, bpz014.
- Lima, A.R.; Pinto, J.; Azevedo, A.I.; Barros-Silva, D.; Jerónimo, C.; Henrique, R.; de Lourdes Bastos, M.; de Pinho, P.G.; Carvalho, M. Identification of a biomarker panel for improvement of prostate cancer diagnosis by volatile metabolic profiling of urine. Br. J. Cancer 2019, 121, 857–868.
- Lima, A.R.; Bastos, M.d.L.; Carvalho, M.; Guedes de Pinho, P. Biomarker Discovery in Human Prostate Cancer: An Update in Metabolomics Studies. Transl. Oncol. 2016, 9, 357–370.
- Silva, C.; Perestrelo, R.; Silva, P.; Tomás, H.; Câmara, J.S. Breast Cancer Metabolomics: From Analytical Platforms to Multivariate Data Analysis. A Review. Metabolites 2019, 9, 102.
- Gao, Q.; Lee, W.Y. Urinary metabolites for urological cancer detection: A review on the application of volatile organic compounds for cancers. Am. J. Clin. Exp. Urol. 2019, 7, 232–248.
- Khalid, T.; Aggio, R.; White, P.; De Lacy Costello, B.; Persad, R.; Al-Kateb, H.; Jones, P.; Probert, C.S.; Ratcliffe, N. Urinary Volatile Organic Compounds for the Detection of Prostate Cancer. PLoS ONE 2015, 10, e0143283.
- Struck-Lewicka, W.; Kordalewska, M.; Bujak, R.; Yumba Mpanga, A.; Markuszewski, M.; Jacyna, J.; Matuszewski, M.; Kaliszan, R.; Markuszewski, M.J. Urine metabolic fingerprinting using LC-MS and GC-MS reveals metabolite changes in prostate cancer: A pilot study. J. Pharm. Biomed. Anal. 2015, 111, 351–361.
- Gao, Q.; Su, X.; Annabi, M.H.; Schreiter, B.R.; Prince, T.; Ackerman, A.; Morgas, S.; Mata, V.; Williams, H.; Lee, W.-Y. Application of Urinary Volatile Organic Compounds (VOCs) for the Diagnosis of Prostate Cancer. Clin. Genitourin. Cancer 2019, 17, 183–190.
- Bax, C.; Taverna, G.; Eusebio, L.; Sironi, S.; Grizzi, F.; Guazzoni, G.; Capelli, L. Innovative Diagnostic Methods for Early Prostate Cancer Detection through Urine Analysis: A Review. Cancers (Basel) 2018, 10, 123.
- da Costa, B.R.B.; De Martinis, B.S. Analysis of urinary VOCs using mass spectrometric methods to diagnose cancer: A review. Clin. Mass Spectrom. 2020, 18, 27–37.
- Tyagi, H.; Daulton, E.; Bannaga, A.S.; Arasaradnam, R.P.; Covington, J.A. Urinary Volatiles and Chemical Characterisation for the Non-Invasive Detection of Prostate and Bladder Cancers. Biosensors (Basel) 2021, 11, 437.
- Lima, A.R.; Pinto, J.; Carvalho-Maia, C.; Jerónimo, C.; Henrique, R.; Bastos, M.d.L.; Carvalho, M.; Guedes de Pinho, P. A Panel of Urinary Volatile Biomarkers for Differential Diagnosis of Prostate Cancer from Other Urological Cancers. Cancers 2020, 12, 2017.
- Capelli, L.; Taverna, G.; Bellini, A.; Eusebio, L.; Buffi, N.; Lazzeri, M.; Guazzoni, G.; Bozzini, G.; Seveso, M.; Mandressi, A.; et al. Application and Uses of Electronic Noses for Clinical Diagnosis on Urine Samples: A Review. Sensors (Basel) 2016, 16, 1708.
More
