Changes in biological properties over several generations, induced by controlling short-term evolutionary processes in the laboratory through selective pressure, and whole-genome re-sequencing, help determine the genetic basis of microorganism's adaptive laboratory evolution (ALE). Due to the versatility of this technique and the imminent urgency for alternatives to petroleum-based strategies, ALE has been actively conducted for several yeasts, primarily using the conventional species Saccharomyces cerevisiae, but also non-conventional yeasts. As a hot topic at the moment since genetically modified organisms are a debatable subject and a global consensus on their employment has not yet been attained, a panoply of new studies employing ALE approaches have emerged and many different applications have been exploited in this context. In the present review, we gathered, for the first time, relevant studies showing the ALE of non-conventional yeast species towards their biotechnological improvement, cataloging them according to the aim of the study, and comparing them considering the species used, the outcome of the experiment, and the employed methodology. This review sheds light on the applicability of ALE as a powerful tool to enhance species features and improve their performance in biotechnology, with emphasis on the non-conventional yeast species, as an alternative or in combination with genome editing approaches.
- yeasts
- adaptive laboratory evolution
- process
1. Introduction
2. Experimental Approaches of Adaptive Laboratory Evolution
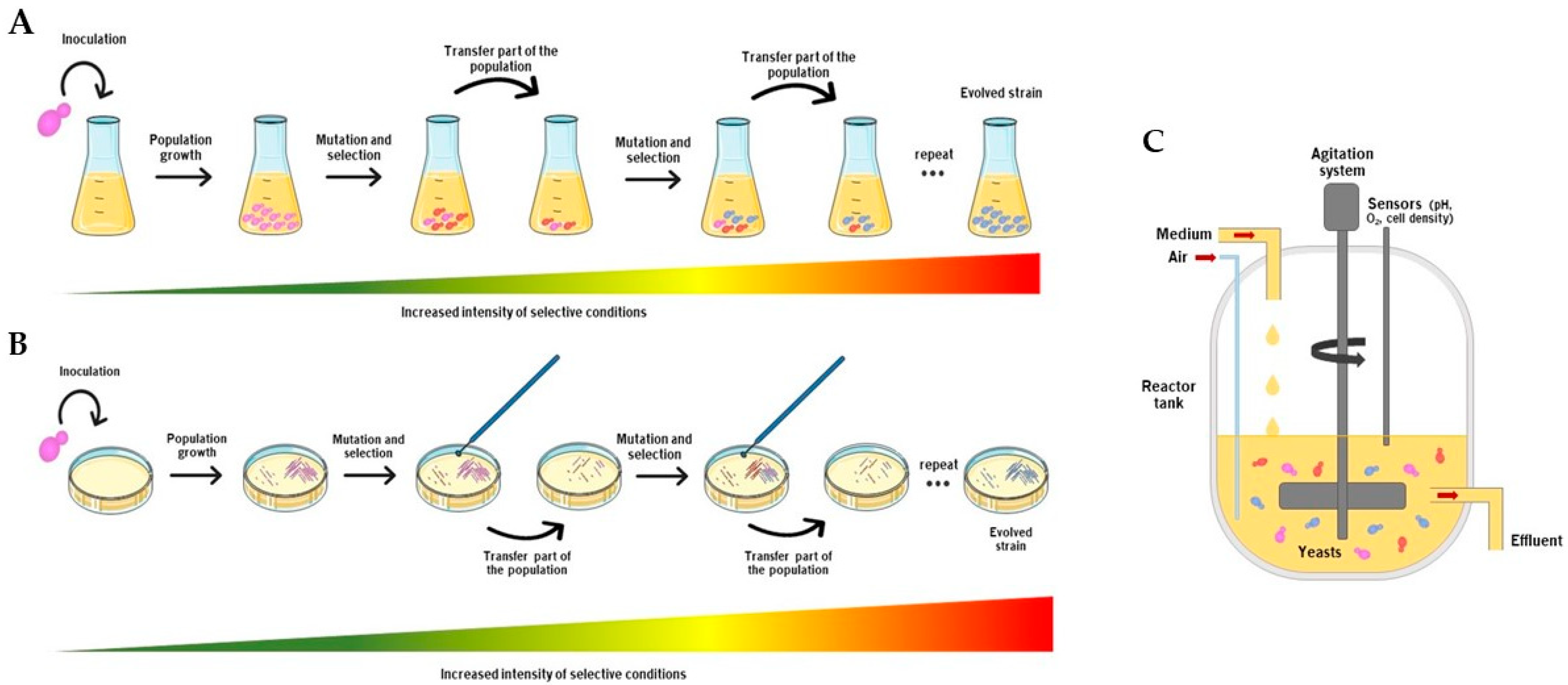
2.1. Serial Batch Cultures
| Main Applications | Advantages | Disadvantages | References | |
|---|---|---|---|---|
| Serial Batch |
|
|
|
[2][3][5][6] |
| Continuous culture |
|
|
|
[2][3][6][7][8][9] |
| Automated methods |
|
|
|
[1][3][6] |
2.2. Continuous Culture
3. Automated Methods for Adaptive LEaboratory Evolution: High-Throughput Adaptive Evolution
References
- Lee, S.; Kim, P. Current status and applications of adaptive laboratory evolution in industrial microorganisms. J. Microbiol. Biotechnol. 2020, 30, 793–803.
- Dragosits, M.; Mattanovich, D. Adaptive laboratory evolution–principles and applications for biotechnology. Microb. Cell Factories 2013, 12, 64.
- Winkler, J.; Reyes, L.H.; Kao, K.C. Adaptive Laboratory Evolution for Strain Engineering. In Systems Metabolic Engineering; Humana Press: Totowa, NJ, USA, 2013; pp. 211–222.
- Choe, D.; Lee, J.H.; Yoo, M.; Hwang, S.; Sung, B.H.; Cho, S.; Palsson, B.; Kim, S.C.; Cho, B.K. Adaptive laboratory evolution of a genome-reduced Escherichia coli. Nat. Commun. 2019, 10, 935.
- Gonzalez, A.; Bell, G. Evolutionary rescue and adaptation to abrupt environmental change depends upon the history of stress. Philos. Trans. R. Soc. B Biol. Sci. 2013, 368, 20120079.
- Sandberg, T.E.; Salazar, M.J.; Weng, L.L.; Palsson, B.Ø.; Feist, A.M. The emergence of adaptive laboratory evolution as an efficient tool for biological discovery and industrial biotechnology. Metab. Eng. 2019, 56, 1–16.
- Gresham, D.; Dunham, M.J. The enduring utility of continuous culturing in experimental evolution. Genomics 2014, 104, 399–405.
- Swamy, K.; Zhou, N. Experimental evolution: Its principles and applications in developing stress-tolerant yeasts. Appl. Microbiol. Biotechnol. 2019, 103, 2067–2077.
- Ekkers, D.M.; Branco dos Santos, F.; Mallon, C.A.; Bruggeman, F.; van Doorn, G.S. The omnistat: A flexible continuous-culture system for prolonged experimental evolution. Methods Ecol. Evol. 2020, 11, 932–942.
- López-Malo, M.; García-Rios, E.; Melgar, B.; Sanchez, M.R.; Dunham, M.J.; Guillamón, J.M. Evolutionary engineering of a wine yeast strain revealed a key role of inositol and mannoprotein metabolism during low-temperature fermentation. BMC Genom. 2015, 16, 537.
- Catrileo, D.; Acuña-Fontecilla, A.; Godoy, L. Adaptive Laboratory Evolution of Native Torulaspora delbrueckii YCPUC10 With Enhanced Ethanol Resistance and Evaluation in Co-inoculated Fermentation. Front. Microbiol. 2020, 11, 1–13.
- Patzschke, A.; Steiger, M.G.; Holz, C.; Lang, C.; Mattanovich, D.; Sauer, M. Enhanced glutathione production by evolutionary engineering of Saccharomyces cerevisiae strains. Biotechnol. J. 2015, 10, 1719–1726.
- Mozhayskiy, V.; Tagkopoulos, I. Microbial evolution in vivo and in silico: Methods and applications. Integr. Biol. 2013, 5, 262–277.
- Horinouchi, T.; Furusawa, C. Understanding metabolic adaptation by using bacterial laboratory evolution and trans-omics analysis. Biophys. Rev. 2020, 12, 677–682.
- McCloskey, D.; Xu, S.; Sandberg, T.E.; Brunk, E.; Hefner, Y.; Szubin, R.; Feist, A.M.; Palsson, B.Ø. Adaptive laboratory evolution resolves energy depletion to maintain high aromatic metabolite phenotypes in Escherichia coli strains lacking the phosphotransferase system. Metab. Eng. 2018, 48, 233–242.
- Vasconcellos, V.M.; Farinas, C.S.; Ximenes, E.; Slininger, P.; Ladisch, M. Adaptive laboratory evolution of nanocellulose-producing bacterium. Biotechnol. Bioeng. 2019, 116, 1923–1933.
- Kurtzman, C.P.; Fell, J.W.; Boekhout, T. The Yeasts: A Taxonomic Study; Elsevier: Amsterdam, The Netherlands, 2011.
- Drumonde-Neves, J.; Fernandes, T.; Lima, T.; Pais, C.; Franco-Duarte, R. Learning from 80 years of studies: A comprehensive catalogue of non-Saccharomyces yeasts associated with viticulture and winemaking. FEMS Yeast Res. 2021, 21, foab017.
- Fernandes, T.; Silva-Sousa, F.; Pereira, F.; Rito, T.; Soares, P.; Franco-Duarte, R.; Sousa, M.J. Biotechnological Importance of Torulaspora delbrueckii: From the Obscurity to the Spotlight. J. Fungi 2021, 7, 712.
- Silva-Sousa, F.; Fernandes, T.; Pereira, F.; Rodrigues, D.; Rito, T.; Camarasa, C.; Franco-Duarte, R.; Sousa, M.J. Torulaspora delbrueckii Phenotypic and Metabolic Profiling towards Its Biotechnological Exploitation. J. Fungi 2022, 8, 569.
- Franco-Duarte, R.; Mendes, I.; Umek, L.; Drumonde-Neves, J.; Zupan, B.; Schuller, D. Computational models reveal genotype–phenotype associations in Saccharomyces cerevisiae. Yeast 2014, 31, 265–277.
- Zheng, Y.; Hong, K.; Wang, B.; Liu, D.; Chen, T.; Wang, Z. Genetic diversity for accelerating microbial adaptive laboratory evolution. ACS Synth. Biol. 2021, 10, 1574–1586.
- Coulon, J.; Husnik, J.I.; Inglis, D.L.; van der Merwe, G.K.; Lonvaud, A.; Erasmus, D.J.; van Vuuren, H.J. Metabolic engineering of Saccharomyces cerevisiae to minimize the production of ethyl carbamate in wine. Am. J. Enol. Vitic. 2006, 57, 113–124.
- Grossmann, M.; Kießling, F.; Singer, J.; Schoeman, H.; Schröder, M.B.; von Wallbrunn, C. Genetically modified wine yeasts and risk assessment studies covering different steps within the wine making process. Ann. Microbiol. 2011, 61, 103–115.
- Husnik, J.I.; Volschenk, H.; Bauer, J.; Colavizza, D.; Luo, Z.; Van Vuuren, H.J. Metabolic engineering of malolactic wine yeast. Metab. Eng. 2006, 8, 315–323.
- DiCarlo, J.E.; Norville, J.E.; Mali, P.; Rios, X.; Aach, J.; Church, G.M. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013, 41, 4336–4343.
- Turnbull, C.; Lillemo, M.; Hvoslef-Eide, T.A. Global Regulation of Genetically Modified Crops Amid the Gene Edited Crop Boom—A Review. Front. Plant Sci. 2013, 12, 258.
- Gonzalez, R.; Tronchoni, J.; Quirós, M.; Morales, P. Genetic Improvement and Genetically Modified Microorganisms. In Wine Safety, Consumer Preference, and Human Health; Springer: Cham, Switzerland, 2016; pp. 71–96.
- Hass, J.W., Jr. The Reverend Dr William Henry Dallinger, F.R.S. (1839–1909). Notes Rec. R. Soc. London. 2000, 54, 53–65.
- Bailey, L.A.; Hatton, D.; Field, R.; Dickson, A.J. Determination of Chinese hamster ovary cell line stability and recombinant antibody expression during long-term culture. Biotechnol. Bioeng. 2012, 109, 2093–2103.
- Kering, K.K.; Zhang, X.; Nyaruaba, R.; Yu, J.; Wei, H. Application of adaptive evolution to improve the stability of bacteriophages during storage. Viruses 2020, 12, 423.
- Garcia-Rios, E.; Lairon-Peris, M.; Muniz-Calvo, S.; Heras, J.M.; Ortiz-Julien, A.; Poirot, P.; Rozès, N.; Querol, A.; Guillamón, J.M. Thermo-adaptive evolution to generate improved Saccharomyces cerevisiae strains for cocoa pulp fermentations. Int. J. Food Microbiol. 2021, 342, 109077.
- Xia, H.; Kang, Y.; Ma, Z.; Hu, C.; Yang, Q.; Zhang, X.; Yang, S.; Dai, J.; Chen, X. Evolutionary and reverse engineering in Saccharomyces cerevisiae reveals a Pdr1p mutation-dependent mechanism for 2-phenylethanol tolerance. Microb. Cell Factories 2022, 21, 269.
- Barten, R.; van Workum, D.J.M.; de Bakker, E.; Risse, J.; Kleisman, M.; Navalho, S.; Smit, S.; Wijffels, R.H.; Nijveen, H.; Barbosa, M.J. Genetic mechanisms underlying increased microalgal thermotolerance, maximal growth rate, and yield on light following adaptive laboratory evolution. BMC Biol. 2022, 20, 242.
- He, Y.; Yin, H.; Dong, J.; Yu, J.; Zhang, L.; Yan, P.; Wan, X.; Hou, X.; Zhao, Y.; Chen, R.; et al. Reduced sensitivity of lager brewing yeast to premature yeast flocculation via adaptive evolution. Food Microbiol. 2022, 106, 104032.
- Voordeckers, K.; Kominek, J.; Das, A.; Espinosa-Cantú, A.; De Maeyer, D.; Arslan, A.; Van Pee, M.; van der Zande, E.; Meert, W.; Yang, Y.; et al. Adaptation to high ethanol reveals complex evolutionary pathways. PLoS Genet. 2015, 11, e1005635.
- Heins, Z.J.; Mancuso, C.P.; Kiriakov, S.; Wong, B.G.; Bashor, C.J.; Khalil, A.S. Designing automated, high-throughput, continuous cell growth experiments using eVOLVER. J. Vis. Exp. 2019, 147, e59652.
- Zhong, Z.; Wong, B.G.; Ravikumar, A.; Arzumanyan, G.A.; Khalil, A.S.; Liu, C.C. Automated Continuous Evolution of Proteins in Vivo. ACS Synth. Biol. 2020, 9, 1270–1276.
- LaCroix, R.A.; Palsson, B.Ø.; Feist, A.M. A model for designing adaptive laboratory evolution experiments. Appl. Environ. Microbiol. 2017, 83, 1–14.
