You're using an outdated browser. Please upgrade to a modern browser for the best experience.
Please note this is a comparison between Version 2 by Sirius Huang and Version 1 by Qiushui He.
Pertussis is a highly contagious respiratory infection caused by Bordetella pertussis bacterium. The mainstay of treatment is macrolide antibiotics that reduce transmissibility, shorten the duration of symptoms and decrease mortality in infants.
- Bordetella pertussis
- pertussis
- whooping cough
- macrolides
- macrolide resistance
1. Introduction
Pertussis, or whooping cough, is a highly contagious respiratory infection caused by Bordetella pertussis, a small Gram-negative rod bacterium. Despite extensive vaccinations, whooping cough is resurging in many countries including USA, UK and China [1]. The disease can manifest as a severe life-threatening illness, especially in unvaccinated young infants. A cornerstone of the clinical management of infants with recent onset of pertussis infection is, in addition to supportive care, antibiotic management by macrolide antibiotics. Macrolide treatment might ameliorate the disease when started early after infection onset, before the appearance of paroxysmal cough [2].
Macrolides (erythromycin (ERY), clarithromycin (CHL) and azithromycin (AZT)] are the first line antimicrobials used to treat pertussis patients. Several studies have shown their efficacy in vitro, and in clinical settings for clearance of B. pertussis [3,4,5,6][3][4][5][6].
The first B. pertussis strain with decreased sensitivity to macrolide antibiotics was detected in Arizona, USA in 1994 [7]. Since then, macrolide resistant B. pertussis has been detected in several countries, although it is rare. However, macrolide resistant B. pertussis has been increasingly reported in China during past decade, raising the concern of its potential transmission to other regions and countries.
2. Pertussis Diagnostics
Pertussis diagnostics can be divided into three main approaches: (1) culture, (2) nucleic acid detection (PCR) and (3) serology. Patient age, vaccination history and onset of the symptoms should be considered when choosing the correct diagnostic method [8]. Culture can be performed up to 2 weeks after the symptoms have appeared, before the bacteria is cleared by the immune defence. Specimen from freshly obtained nasopharyngeal (NP) samples should be cultured on Regan-Lowe (RL, charcoal) or Bordet-Gengou (BG, blood) agar containing cephalexin. Suspected B. pertussis specific colonies are further cultured on RL/BG agar (without cephalexin), and identified with e.g., slide agglutination test with specific anti-B. pertussis and anti-B. parapertussis sera or MALDI-TOF [8,9,10][8][9][10]. Specific nucleic acid identification (targeting IS481/ptxp) with PCR requires only a small amount of DNA for detection and identification of the bacterium and is therefore far more sensitive than culture. Furthermore, it can be used even three to four weeks after the onset of symptoms. Therefore, PCR-based approaches are more widely used than culture, especially with infants and small children. For school children and adults, serology is commonly used as there is less interference in antibodies induced from previous vaccinations and the only symptom may have been a prolonged cough (>3–4 weeks, culture nor PCR can be used). Serological diagnosis should be made based on the measurement of serum IgG antibodies against pertussis toxin [11]. Furthermore, laboratory confirmation of B. pertussis from clinical samples is needed before antimicrobial susceptibility testing (AST) is performed.3. Epidemiology
The first macrolide resistant B. pertussis strain was identified in a 2-month-old infant from Yuma, Arizona, US in 1994 [7]. The isolate was highly resistant to erythromycin with a minimum inhibitory concentration (MIC) > 64 µg/mL. However, the origin of this isolate was not known. Breakpoints to detect antimicrobial resistance of clinical B. pertussis isolates were not standardized but the reported resistant strains had MICs of >256 µg/mL with erythromycin (ERY) and clarithromycin (CHL) by Etest method suggesting macrolide resistance. Concurrently, seven additional B. pertussis isolates from the same area were tested, but macrolide resistance was not detected in these cases. In a review of 47 B. pertussis isolates from children in Utah, US, in 1985–1997, one isolate from January 1997 was resistant against erythromycin [12]. Since the first appearance of macrolide-resistant B. pertussis, macrolide susceptibility has been tested in thousands of cultured isolates all over the world (Table 1, Figure 1). In a study of 1030 isolates collected from various parts of the the US, five (0.5%) isolates were erythromycin resistant. Four out of five isolates were from Arizona (1994–1995) and one from Georgia (1995). All isolates initially showed the growth inhibition of B. pertussis by disc diffusion method, but after 5–7 days of incubation, novel bacterial colonies appeared on the plate inside the growth inhibition area, demonstrating heterogeneous phenotype [13]. In a review of 38 B. pertussis isolates from France in 2003, none of them were resistant to erythromycin [14]. However, nine years later in 2012, the first patient in Europe with macrolide-resistant B. pertussis was diagnosed in Lyon, France [15]. A three-week-old neonate with severe pertussis was treated repeatedly with macrolides before the detection of the resistant isolate. Of the three serial isolates from the patient, the first two were sensitive, but the third one turned to be resistant, suggesting that the B. pertussis isolate acquired the mutation leading to macrolide resistance during the macrolide treatment. Sporadic cases of macrolide-resistant B. pertussis isolates were also reported from Iran in 2009 [16].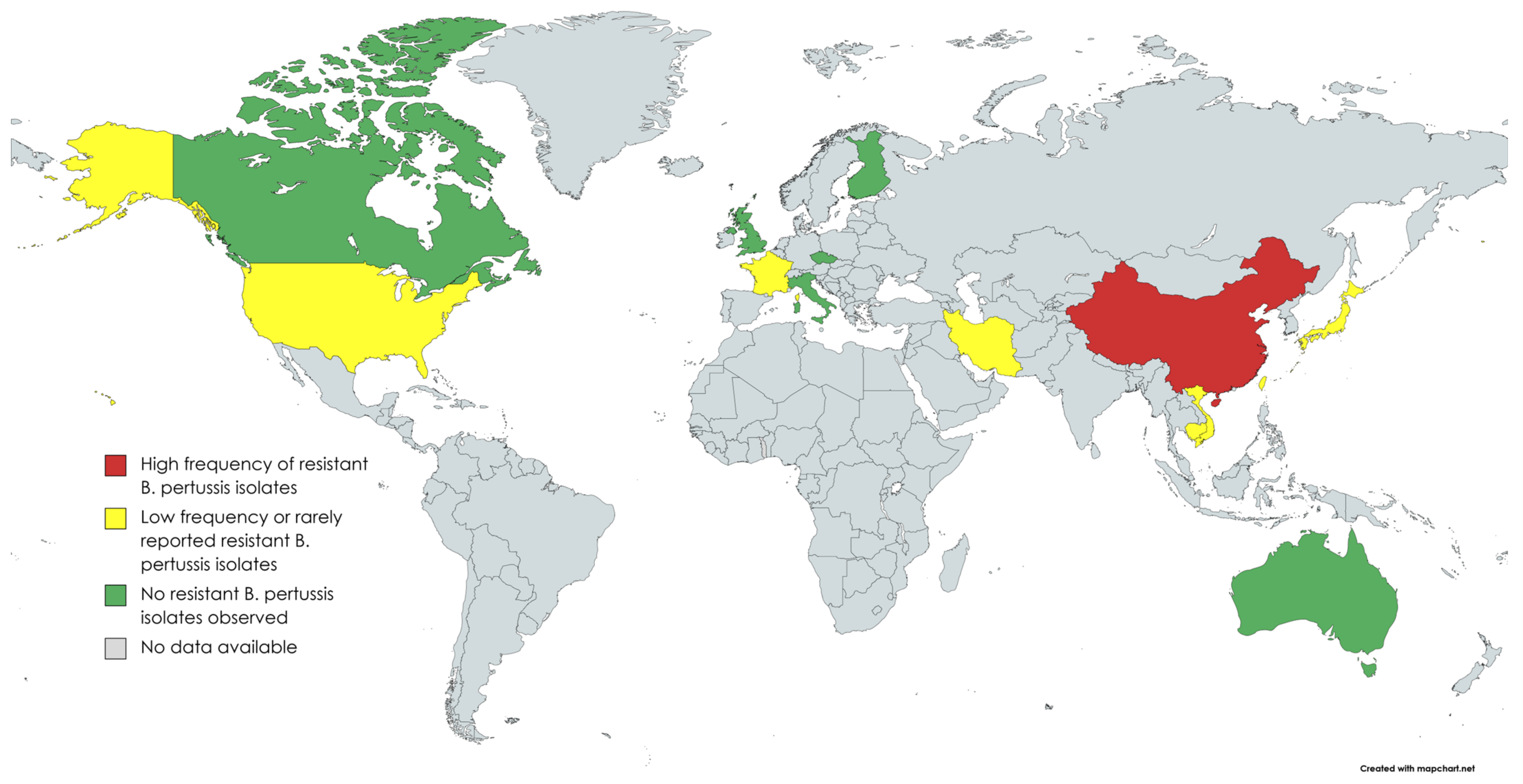
Figure 1.
Countries where
B. pertussis
antimicrobial susceptibility studies have been performed (created with MapChart).
Table 1.
Global frequencies of macrolide-resistant
Bordetella pertussis
.
| Country | Region/City | Year | Resistant Isolates Identified (Frequency %) |
Reference |
|---|---|---|---|---|
| Australia | New South Wales, Perth | 1971–2010 | 0/120 (0.0) | [24,25][24][25] |
| Cambodia | Whole country | 2017–2020 | 1/71 (1.4) | [19] |
| Canada | Ontario | 2011–2013 | 0/275 (0.0) | [26] |
| China | Xi’an | 2012–2020 | 274/299 (91.6) | [27,28,29,30,31][27][28][29][30][31] |
| Shandong | 2011 | 2/2 (100.0) | [21] | |
| Northern | 1970–2014 ** | 91/124 ** (91.9) | [22] | |
| Shanghai | 2016–2017 | 81/141 (57.5) | [32] | |
| Zhejiang | 2016–2020 | 271/381 (71.1) | [33,34,35][33][34][35] | |
| Beijing, Jinan, Nanjing, Shenzhen | 2014–2016 | 292/335 (87.2) | [36] | |
| Midwest | 2012–2015 | 163/167 (97.6) | [37] | |
| Whole country | 1950–2018 | 316/388 (81.4) | [23] | |
| Hunan | 2017–2018 | 27/55 (49.1) | [38] | |
| Shenzhen | 2015–2017 | 51/105 (48.6) | [39] | |
| Whole country | 2017–2019 | 265/311 (85.2) | [40] | |
| Czech republic | Whole country | 1967–2015 | 0/135 (0.0) | [41] |
| Finland | Whole country | 2006–2017 | 0/148 (0.0) | [42] |
| France | Bordeaux & Lyon | 2003 and 2012 | 1/41 (2.4) | [10,11][10][11] |
| Iran | Whole country | 2009–2010 | 2/11 (18.2) | [16,43][16][43] |
| Italy | Rome | 2012–2015 | 0/18 (0.0) | [44] |
| Japan | Whole country | 2017–2019 | 1/33 (3.0) | [17,19][17][19] |
| Taiwan | Whole country | 2003–2007 | 2/76 (2.6) | [19,23][19][23] |
| United Kingdom | Whole country | 2001–2009 | 0/582 (0.0) | [45] |
| United States | Colorado, Maryland, Oklahoma, Wisconsin | 1986 | 0/75 (0.0) | [46] |
| Arizona—Yuma County | 1994 | 1/1 (100.0) | [47] | |
| Utah | 1985–1997 | 1/47 (2.1) | [12] | |
| Northern California | 1998–1999 | 0/36 (0.0) | [48] | |
| Phoenix, Oakland *, San Diego | N/A *** | 1/48 (2.1) | [49] | |
| California, New York, Minnesota, Massachusetts, Illinois, Arizona, Georgia | 1994–2000 | 5/1030 **** (0.5) | [13] | |
| Minnesota | 1997–1999 | 1/8 (12.5) | [50] | |
| Vietnam | Hanoi, Ha Nam, Thai Binh | 2016–2020 | 24/184 (13.0) | [18,19][18][19] |
* Hill et al. included a control B. pertussis strain, resistant to macrolides. This strain has been isolated in Oakland but not officially published elsewhere. ** Divided into three time periods: 1970s, 2000–2008 and 2013–2014. All isolates (N = 25) collected in 1970–2008 were macrolide sensitive. *** N/A = Not available. **** Notified 5 to 7 days after incubation. Four from Arizona, one from Georgia.
Until recently, macrolide resistance in B. pertussis in China has been associated almost exclusively with the ptxP1 lineage of the bacterium [22,27,29,30,31,32,37][22][27][29][30][31][32][37]. However, a recent cross-sectional study describes two ptxP3 isolates from eastern China that had acquired the A2047G mutation in their 23S rRNA gene [40]. The ptxP3 lineage is currently the dominating B. pertussis circulating in most of the high-income countries that have switched to acellular pertussis vaccine in recent decades [51,52][51][52]. It has been hypothesized that the replacement of the whole-cell pertussis vaccine with co-purified acellular pertussis vaccine in the national immunization programme, the liberal use of macrolides in children with respiratory infections, and high population densities could have contributed to the effective spread of macrolide-resistant B. pertussis in China [53].
4. Mechanisms behind Macrolide Resistance in B. pertussis
Macrolide resistance can be caused by three distinct mechanisms. The most common mechanism, including for B. pertussis, is the A2047G single nucleotide polymorphism (SNP) in the 23S rRNA gene within the domain V [15,28,50][15][28][50]. This is equal to a SNP in position A2058G in E. coli and A2064G in M. pneumoniae [54,55][54][55]. The A2047G mutation affects the macrolide binding site in the 23S rRNA component of the 50S ribosomal subunit and prevents macrolides to inhibit the peptide elongation [50]. There are three copies of this gene in the B. pertussis genome. Bartkus et al. showed that the A2047G SNP can be found in one or more of the copies. They suggested that this mutation needs at least two copies for resistance [50]. However, many studies have shown that in most cases, all three copies are mutated among the macrolide-resistant B. pertussis strains [15,27,37][15][27][37]. The second possible cause is the acquisition of the ERY-resistant methylase (erm) gene, which leads to addition of methyl group in the 23S rRNA to block the ERY binding site [37,50][37][50]. However, B. pertussis do not possess this gene, which is also shown in a novel study in which 167 clinical isolates were screened to identify the possible inclusion of this gene. However, none of the strains carried such a gene [37]. So far, no studies have found this mechanism to be the cause of macrolide resistance in B. pertussis. The third proposed mechanism is the expression of MexAB-OprM efflux pump (regulated by the mexAB-oprM operon), which helps the bacteria to regulate the uptake of macrolides. This mechanism excretes macrolide molecules out of the bacterial cell. The mechanism has been well-described and has been shown to cause resistance against many antimicrobial agents, including macrolides, in Pseudomonas aeruginosa [56]. Lately, Fong et al. described the expression of the mexAB-oprM operon within macrolide-resistant Bordetella parapertussis. Furthermore, they showed upregulation of the mexAB-oprM when B. parapertussis was grown in 256 mg/mL of ERY. As no other mechanism was found to cause the resistance, they speculated on the potential effect of this mechanism to cause the resistance. However, they also showed that this operon was not functional in B. pertussis due to deletions in mexA and oprM genes [57]. Whether there will be B. pertussis with functional mexAB-oprM operon remains to be seen. There have only been two reports (Iran and China) where the A2047G SNP has not been the mechanism behind the macrolide resistance in B. pertussis [22,43][22][43]. However, these two studies did not perform erm gene or mexAB-oprM operon identification, and the reason for the resistance remains unknown. In the study by Mirzaei et al., the macrolide-resistant isolate was resistant to ERY/CHL but not to AZT [43]. Therefore, the presence of erm could be the cause of the resistance in these studies and would be the first one detected among macrolide-resistant B. pertussis.References
- Yeung, K.H.T.; Duclos, P.; Nelson, E.A.S.; Hutubessy, R.C.W. An Update of the Global Burden of Pertussis in Children Younger than 5 Years: A Modelling Study. Lancet Infect. Dis. 2017, 17, 974–980.
- Kimberlin, D.W.; Barnett, E.D.; Lynfield, R.; Sawyer, M.H. (Eds.) Pertussis (Whooping Cough). In Red Book: 2021 Report of the Committee on Infectious Diseases; American Academy of Pediatrics (AAP): Itasca, IL, USA, 2021; pp. 578–589. ISBN 978-1-61002-521-8.
- Bass, J.W.; Crast, F.W.; Kotheimer, J.B.; Mitchell, I.A. Susceptibility of Bordetella Pertussis to Nine Antimicrobial Agents. Am. J. Dis. Child. 1969, 117, 276–280.
- Hardy, D.J.; Hensey, D.M.; Beyer, J.M.; Vojtko, C.; McDonald, E.J.; Fernandes, P.B. Comparative in Vitro Activities of New 14-, 15-, and 16-Membered Macrolides. Antimicrob. Agents Chemother. 1988, 32, 1710–1719.
- Tiwari, T.; Murphy, T.V.; Moran, J. Recommended Antimicrobial Agents for the Treatment and Postexposure Prophylaxis of Pertussis: 2005 CDC Guidelines. MMWR Recomm. Rep. 2005, 54, 1–16.
- Carbonetti, N.H. Bordetella Pertussis: New Concepts in Pathogenesis and Treatment. Curr. Opin. Infect. Dis. 2016, 29, 287–294.
- Lewis, K.; Saubolle, M.A.; Tenover, F.C.; Rudinsky, M.F.; Barbour, S.D.; Cherry, J.D. Pertussis Caused by an Erythromycin-Resistant Strain of Bordetella Pertussis. Pediatr. Infect. Dis. J. 1995, 14, 388–391.
- Wirsing Von König, C.H. Pertussis Diagnostics: Overview and Impact of Immunization. Expert Rev. Vaccines 2014, 13, 1167–1174.
- Zintgraff, J.; Irazu, L.; Lara, C.S.; Rodriguez, M.; Santos, M. The Classical Bordetella Species and MALDI-TOF Technology: A Brief Experience. J. Med. Microbiol. 2018, 67, 1737–1742.
- World Health Organisation (WHO). Laboratory Manual for the Diagnosis of Whooping Cough Caused by Bordetella Pertussis/Bordetella Parapertussis. Update 2014; World Health Organisation (WHO): Geneva, Switzerland, 2014.
- Guiso, N.; Liese, J.; Plotkin, S. The Global Pertussis Initiative: Meeting Report from the Fourth Regional Roundtable Meeting, France, April 14–15, 2010. Hum. Vaccin. 2011, 7, 481–488.
- Korgenski, E.K.; Daly, J.A. Surveillance and Detection of Erythromycin Resistance in Bordetella Pertussis Isolates Recovered from a Pediatric Population in the Intermountain West Region of the United States. J. Clin. Microbiol. 1997, 35, 2989–2991.
- Wilson, K.E.; Cassiday, P.K.; Popovic, T.; Sanden, G.N. Bordetella Pertussis Isolates with a Heterogeneous Phenotype for Erythromycin Resistance. J. Clin. Microbiol. 2002, 40, 2942–2944.
- Bourgeois, N.; Ghnassia, J.C.; Doucet-Populaire, F. In Vitro Activity of Fluoroquinolones against Erythromycin-Susceptible and -Resistant Bordetella Pertussis. J. Antimicrob. Chemother. 2003, 51, 742–743.
- Guillot, S.; Descours, G.; Gillet, Y.; Etienne, J.; Floret, D.; Guiso, N. Macrolide-Resistant Bordetella Pertussis Infection in Newborn Girl, France. Emerg. Infect. Dis. 2012, 18, 966.
- Shahcheraghi, F.; Lotfi, M.N.; Nikbin, V.S.; Shooraj, F.; Azizian, R.; Parzadeh, M.; Torkaman, M.R.A.; Zahraei, S.M. The First Macrolide-Resistant Bordetella Pertussis Strains Isolated From Iranian Patients. Jundishapur J. Microbiol. 2014, 7, 10880.
- Yamaguchi, T.; Kawasaki, Y.; Katsukawa, C.; Kawahara, R.; Kawatsu, K. The First Report of Macrolide-Resistant Bordetella Pertussis Isolation in Japan. Jpn. J. Infect. Dis. 2020, 73, 361–362.
- Kamachi, K.; Duong, H.T.; Dang, A.D.; Do, H.T.; Koide, K.; Otsuka, N.; Shibayama, K.; Hoang, H.T.T. Macrolide-Resistant Bordetella Pertussis, Vietnam, 2016−2017. Emerg. Infect. Dis. 2020, 26, 2511–2513.
- Koide, K.; Yao, S.; Chiang, C.; Thuy, P.T.B.; Nga, D.T.T.; Huong, D.T.; Dien, T.M.; Vichit, O.; Vutthikol, Y.; Sovannara, S.; et al. Genotyping and Macrolide-Resistant Mutation of Bordetella Pertussis in East and South-East Asia. J. Glob. Antimicrob. Resist. 2022, 31, 263–269.
- Koide, K.; Yamaguchi, T.; Katsukawa, C.; Otsuka, N.; Kenri, T.; Kamachi, K. Complete Genome Sequence of a Macrolide-Resistant Bordetella Pertussis Isolated in Japan. Microbiol. Resour. Announc. 2022, 11, e00718-22.
- Zhang, Q.; Li, M.; Wang, L.; Xin, T.; He, Q. High-Resolution Melting Analysis for the Detection of Two Erythromycin-Resistant Bordetella Pertussis Strains Carried by Healthy Schoolchildren in China. Clin. Microbiol. Infect. 2013, 19, E260–E262.
- Yang, Y.; Yao, K.; Ma, X.; Shi, W.; Yuan, L.; Yang, Y. Variation in Bordetella Pertussis Susceptibility to Erythromycin and Virulence-Related Genotype Changes in China (1970–2014). PLoS ONE 2015, 10, e0138941.
- Yao, K.; Deng, J.; Ma, X.; Dai, W.; Chen, Q.; Zhou, K.; Ye, J.; Shi, W.; Wang, H.; Li, D.; et al. The Epidemic of Erythromycin-Resistant Bordetella Pertussis with Limited Genome Variation Associated with Pertussis Resurgence in China. Expert Rev. Vaccines 2020, 19, 1093–1099.
- Sintchenko, V.; Brown, M.; Gilbert, G.L. Is Bordetella Pertussis Susceptibility to Erythromycin Changing? MIC Trends among Australian Isolates 1971–2006. J. Antimicrob. Chemother. 2007, 60, 1178–1179.
- Dorji, D.; Graham, R.M.; Richmond, P.; Keil, A.; Mukkur, T.K. Biofilm Forming Potential and Antimicrobial Susceptibility of Newly Emerged Western Australian Bordetella Pertussis Clinical Isolates. Biofouling 2016, 32, 1141–1152.
- Marchand-Austin, A.; Memari, N.; Patel, S.N.; Tang, P.; Deeks, S.L.; Jamieson, F.B.; Crowcroft, N.S.; Farrell, D.J. Surveillance of Antimicrobial Resistance in Contemporary Clinical Isolates of Bordetella Pertussis in Ontario, Canada. Int. J. Antimicrob. Agents 2014, 44, 82–84.
- Wang, Z.; Li, Y.; Hou, T.; Liu, X.; Liu, Y.; Yu, T.; Chen, Z.; Gao, Y.; Li, H.; He, Q. Appearance of Macrolide-Resistant Bordetella Pertussis Strains in China. Antimicrob. Agents Chemother. 2013, 57, 5193–5194.
- Wang, Z.; Cui, Z.; Li, Y.; Hou, T.; Liu, X.; Xi, Y.; Liu, Y.; Li, H.; He, Q. High Prevalence of Erythromycin-Resistant Bordetella Pertussis in Xi’an, China. Clin. Microbiol. Infect. 2014, 20, O825–O830.
- Wang, Z.; Luan, Y.; Du, Q.; Shu, C.; Peng, X.; Wei, H.; Hou, T.; Liu, Y.; Liu, X.; Li, Y. The Global Prevalence PtxP3 Lineage of Bordetella Pertussis Was Rare in Young Children with the Co-Purified APV Vaccination: A 5 Years Retrospective Study. BMC Infect. Dis. 2020, 20, 615.
- Liu, X.; Wang, Z.; Zhang, J.; Li, F.; Luan, Y.; Li, H.; Li, Y.; He, Q. Pertussis Outbreak in a Primary School in China: Infection and Transmission of the Macrolide-Resistant Bordetella Pertussis. Pediatr. Infect. Dis. J. 2018, 37, E145–E148.
- Zhang, J.; Zhang, D.; Wang, X.; Wei, X.; Li, H. Macrolide Susceptibility and Molecular Characteristics of Bordetella Pertussis. J. Int. Med. Res. 2022, 50, 03000605221078782.
- Fu, P.; Wang, C.; Tian, H.; Kang, Z.; Zeng, M. Bordetella Pertussis Infection in Infants and Young Children in Shanghai, China, 2016-2017: Clinical Features, Genotype Variations of Antigenic Genes and Macrolides Resistance. Pediatr. Infect. Dis. J. 2019, 38, 370–376.
- Hua, C.Z.; Wang, H.J.; Zhang, Z.; Tao, X.F.; Li, J.P.; Mi, Y.M.; Tang, L.F.; Chen, Z.M. In Vitro Activity and Clinical Efficacy of Macrolides, Cefoperazone-Sulbactam and Piperacillin/Piperacillin-Tazobactam against Bordetella Pertussis and the Clinical Manifestations in Pertussis Patients Due to These Isolates: A Single-Centre Study in Zheji. J. Glob. Antimicrob. Resist. 2019, 18, 47–51.
- Mi, Y.M.; Hua, C.Z.; Fang, C.; Liu, J.J.; Xie, Y.P.; Lin, L.N.; Wang, G.L. Effect of Macrolides and β-Lactams on Clearance of Bordetella Pertussis in the Nasopharynx in Children with Whooping Cough. Pediatr. Infect. Dis. J. 2021, 40, 87–90.
- Lin, L.-N.; Zhou, J.-S.; Hua, C.-Z.; Bai, G.-N.; Mi, Y.-M.; Zhou, M.-M. Epidemiological and Clinical Characteristics of Pertussis in Children and Their Close Contacts in Households: A Cross-Sectional Survey in Zhejiang Province, China. Front. Pediatr. 2022, 10, 1442.
- Li, L.; Deng, J.; Ma, X.; Zhou, K.; Meng, Q.; Yuan, L.; Shi, W.; Wang, Q.; Li, Y.; Yao, K. High Prevalence of Macrolide-Resistant Bordetella Pertussis and PtxP1 Genotype, Mainland China, 2014–2016. Emerg. Infect. Dis. 2019, 25, 2205–2214.
- Xu, Z.; Wang, Z.; Luan, Y.; Li, Y.; Liu, X.; Peng, X.; Octavia, S.; Payne, M.; Lan, R. Genomic Epidemiology of Erythromycin-Resistant Bordetella Pertussis in China. Emerg. Microbes Infect. 2019, 8, 461–470.
- Lin, X.J.; Zou, J.; Yao, K.; Li, L.; Zhong, L. Analysis of Antibiotic Sensitivity and Resistance Genes of Bordetella Pertussis in Chinese Children. Medicine 2021, 100, e24090.
- Tian, S.; Hu, N.; Lou, J.; Chen, K.; Kang, X.; Xiang, Z.; Chen, H.; Wang, D.; Liu, N.; Liu, D.; et al. Characteristics of COVID-19 Infection in Beijing. J. Infect. 2020, 80, 401–406.
- Wu, X.; Du, Q.; Li, D.; Yuan, L.; Meng, Q.; Fu, Z.; Xu, H.; Yao, K.; Zhao, R. A Cross-Sectional Study Revealing the Emergence of Erythromycin-Resistant Bordetella Pertussis Carrying PtxP3 Alleles in China. Front. Microbiol. 2022, 13, 2504.
- Jakubů, V.; Zavadilová, J.; Fabiánová, K.; Urbášková, P. Trends in the Minimum Inhibitory Concentrations of Erythromycin, Clarithromycin, Azithromycin, Ciprofloxacin, and Trimethoprim/Sulfamethoxazole for Strains of Bordetella Pertussis Isolated in the Czech Republic in 1967–2015. Cent. Eur. J. Public Health 2017, 25, 282–286.
- Lönnqvist, E.; Barkoff, A.M.; Mertsola, J.; He, Q. Antimicrobial Susceptibility Testing of Finnish Bordetella Pertussis Isolates Collected during 2006–2017. J. Glob. Antimicrob. Resist. 2018, 14, 12–16.
- Mirzaei, B.; Bameri, Z.; Babaei, R.; Shahcheraghi, F. Isolation of High Level Macrolide Resistant Bordetella Pertussis without Transition Mutation at Domain V in Iran. Jundishapur J. Microbiol. 2015, 8, 18190.
- Stefanelli, P.; Buttinelli, G.; Vacca, P.; Tozzi, A.E.; Midulla, F.; Carsetti, R.; Fedele, G.; Villani, A.; Concato, C.; Carannante, A.; et al. Severe Pertussis Infection in Infants Less than 6 Months of Age: Clinical Manifestations and Molecular Characterization. Hum. Vaccines Immunother. 2017, 13, 1073–1077.
- Fry, N.K.; Duncan, J.; Vaghji, L.; George, R.C.; Harrison, T.G. Antimicrobial Susceptibility Testing of Historical and Recent Clinical Isolates of Bordetella Pertussis in the United Kingdom Using the Etest Method. Eur. J. Clin. Microbiol. Infect. Dis. 2010, 29, 1183–1185.
- Kurzynski, T.A.; Boehm, D.M.; Rott-Petri, J.A.; Schell, R.F.; Allison, P.E. Antimicrobial Susceptibilities of Bordetella Species Isolated in a Multicenter Pertussis Surveillance Project. Antimicrob. Agents Chemother. 1988, 32, 137–140.
- Centers for Disease Control and Prevention (CDC). Erythromycin-Resistant Bordetella Pertussis—Yuma County, Arizona, May–October 1994. MMWR Morb. Mortal. Wkly. Rep. 1994, 43, 807–810.
- Gordon, K.A.; Fusco, J.; Biedenbach, D.J.; Pfaller, M.A.; Jones, R.N. Antimicrobial Susceptibility Testing of Clinical Isolates of Bordetella Pertussis from Northern California: Report from the SENTRY Antimicrobial Surveillance Program. Antimicrob. Agents Chemother. 2001, 45, 3599–3600.
- Hill, B.C.; Baker, C.N.; Tenover, F.C. A Simplified Method for Testing Bordetella Pertussisfor Resistance to Erythromycin and Other Antimicrobial Agents. J. Clin. Microbiol. 2000, 38, 1151–1155.
- Bartkus, J.M.; Juni, B.A.; Ehresmann, K.; Miller, C.A.; Sanden, G.N.; Cassiday, P.K.; Saubolle, M.; Lee, B.; Long, J.; Harrison, A.R.; et al. Identification of a Mutation Associated with Erythromycin Resistance in Bordetella Pertussis: Implications for Surveillance of Antimicrobial Resistance. J. Clin. Microbiol. 2003, 41, 1167–1172.
- Barkoff, A.M.; Mertsola, J.; Pierard, D.; Dalby, T.; Hoegh, S.V.; Guillot, S.; Stefanelli, P.; Van Gent, M.; Berbers, G.; Vestrheim, D.F.; et al. Surveillance of Circulating Bordetella Pertussis Strains in Europe during 1998 to 2015. J. Clin. Microbiol. 2018, 56, e01998-17.
- Bowden, K.E.; Williams, M.M.; Cassiday, P.K.; Milton, A.; Pawloski, L.; Harrison, M.; Martin, S.W.; Meyer, S.; Qin, X.; DeBolt, C.; et al. Molecular Epidemiology of the Pertussis Epidemic in Washington State in 2012. J. Clin. Microbiol. 2014, 52, 3549–3557.
- Feng, Y.; Chiu, C.H.; Heininger, U.; Hozbor, D.F.; Tan, T.Q.; von König, C.H.W. Emerging Macrolide Resistance in Bordetella Pertussis in Mainland China: Findings and Warning from the Global Pertussis Initiative. Lancet Reg. Health West. Pac. 2021, 8, 100098.
- Morozumi, M.; Hasegawa, K.; Kobayashi, R.; Inoue, N.; Iwata, S.; Kuroki, H.; Kawamura, N.; Nakayama, E.; Tajima, T.; Shimizu, K.; et al. Emergence of Macrolide-Resistant Mycoplasma Pneumoniae with a 23S RRNA Gene Mutation. Antimicrob. Agents Chemother. 2005, 49, 2302–2306.
- Weisblum, B. Erythromycin Resistance by Ribosome Modification. Antimicrob. Agents Chemother. 1995, 39, 577–585.
- Pesingi, P.V.; Singh, B.R.; Pesingi, P.K.; Bhardwaj, M.; Singh, S.V.; Kumawat, M.; Sinha, D.K.; Gandham, R.K. MexAB-OprM Efflux Pump of Pseudomonas Aeruginosa Offers Resistance to Carvacrol: A Herbal Antimicrobial Agent. Front. Microbiol. 2019, 10, 2664.
- Fong, W.; Timms, V.; Sim, E.; Pey, K.; Nguyen, T.; Sintchenko, V. Genomic and Transcriptomic Variation in Bordetella spp. Following Induction of Erythromycin Resistance. J. Antimicrob. Chemother. 2022, 77, 3016–3025.
More
