Harnessing the immune system for cancer therapy has shown success, however the response to immunotherapy has been limited. Deciphering the immunopeptidome repertoire of cancer cells is crucial for identifying neoantigens. To date the emphasis has been on mutations. However, there is more to neoantigens than mutations. Thus, there is a need to identify other types of neoantigens that are commonly expressed in a cancer type that are presented by MHC class I and class II, to induce a cytotoxic CD8+ T and CD4+ T response, respectively. The immunopeptidome encompasses protein post-translation modifications (PTMs), which are overlooked by genome or transcriptome profiling. This rentryview covers how the immunopeptidome can yield novel cancer-specific antigens, focusing on PTMs and their applications.
- immunopeptidome
- PTM
- immunotherapy
- cancer vaccine
1. Introduction
2. The Immunopeptidome as a Source of Different Types of Neoantigens
The immunoediting concept has been critical for our understanding of the mechanisms through which the immune system responds to cancer and how tumor cells can evade the immune response [24]. A key factor in the immune response is the recognition of tumor antigens. T cells, through their TCRs, can interact with the myriad of peptides bound to MHC, sorting out self from non-self. Non-canonical tumor antigens, derived from sequences outside of exons or by alternate protein-processing mechanisms, are of increasing interest for immunotherapy [25]. PTMs are mediated by multiple enzymes, some of which may be dysregulated in tumor cells, rendering them potentially tumor specific. Post-translationally modified proteins undergo processing through the proteasome, resulting in peptides that bind to MHC-I for endogenous proteins or MHC-II for exogenous proteins [26]. Dendritic cells (DCs) are antigen-presenting cells (APCs) in cancer that are essential for T and B cell responses via immunopeptides and native protein presentation, respectively [27][28]. PTMs that are restricted to tumor cells have potential as a source of immunopeptides for immunotherapy.3. Post-Translational Modifications as a Source of Tumor Antigens
Whereas a multitude of PTMs are known to occur, most have not been previously investigated in cancer. Nanoscale liquid chromatography coupled mass spectrometry (nanoLC-MS) has contributed significantly to the identification of PTMs in the immunopeptidome through matching the peptide parent mass (MS1) and the fragment mass (MS2) to sequences in the human genome database, allowing for mass shift due to modified amino acids (e.g., +0.984 Da on Arg) for citrullination; (+97.976 Da on Ser, Thr, and Tyr) for phosphorylation; and (+203.079 Da on Ser and Thr) for O-GlcNAc. PTMs that have been identified in the immunopeptidome with demonstrated immunogenicity in cancer (Figure 1).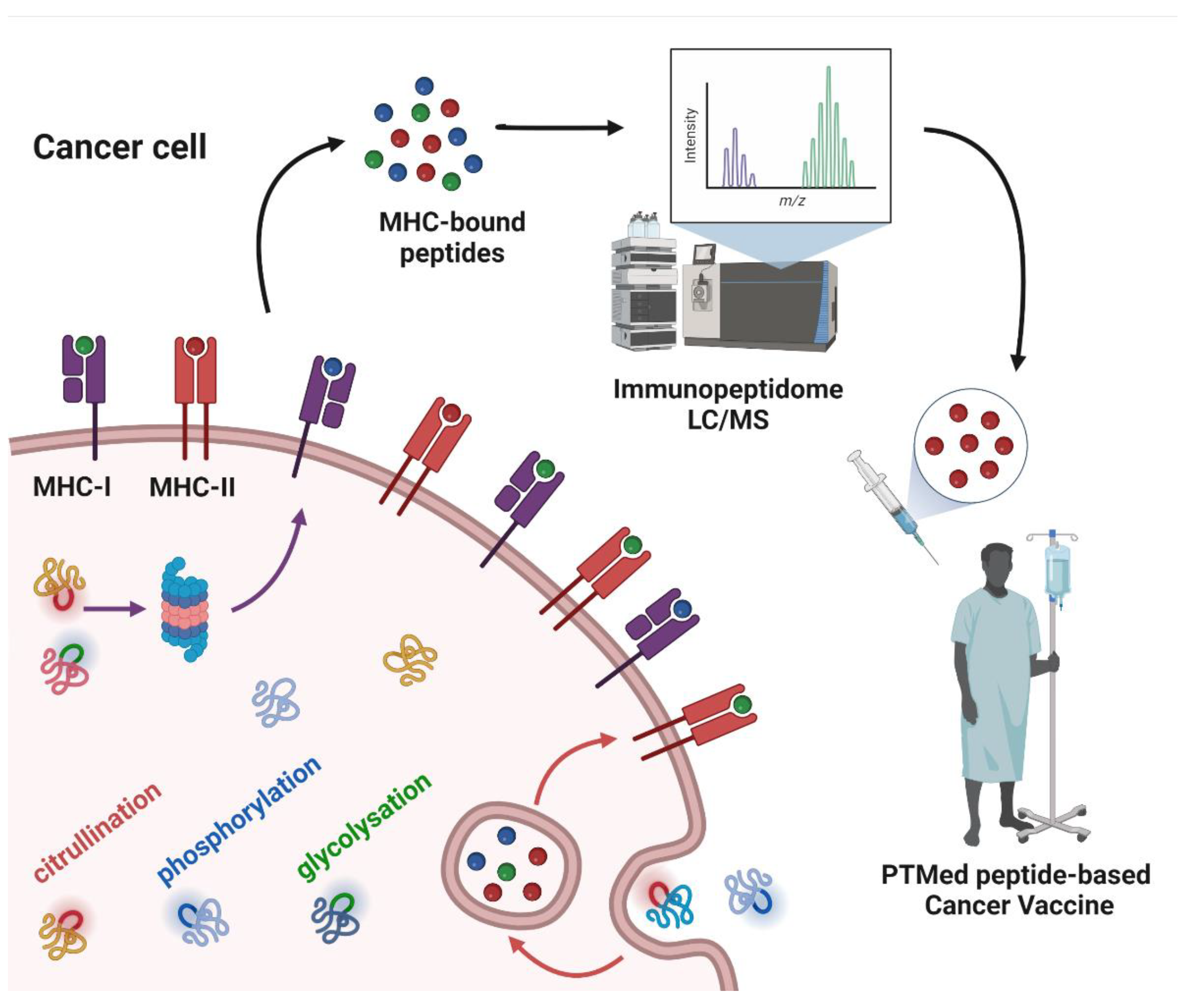
4. Peptide PTMs as a Source of Cancer Vaccines
| Post-Translational Modification |
Protein | Cancer Type | Immunotherapy | MHC Class | Reference |
|---|---|---|---|---|---|
| Citrullination | ENO1 | SKCM, PAAD, LUAD, OV | Vaccine | II | [18][30][31][32] |
| VIM | SKCM, LUAD, PAAD, OV | Vaccine | II | [29][32] | |
| MMP21 | SKCM | Vaccine | II | [33] | |
| GRI | SKCM | Vaccine | II | [33] | |
| Cp450 | SKCM | Vaccine | II | [33] | |
| NPM | SKCM, LUAD | Vaccine | II | [34] | |
| Phosphorylation | ISR2 | SKCM | Vaccine, ACT | I | [35][36] |
| BCAR | SKCM | Vaccine | I | [36] | |
| CDC25b | SKCM | ACT | I | [35] | |
| Glycosylation | MUC1 | BRCA, PRAD | Vaccine, DCTher | I, II | [37][38][39][40] |
| MUC4 | NA | Vaccine | II | [41] | |
| PHOX2B | Neuroblastoma | CAR T cell | I | [42] |
References
- Abbott, M.; Ustoyev, Y. Cancer and the Immune System: The History and Background of Immunotherapy. Semin. Oncol. Nurs. 2019, 35, 150923.
- Kote, S.; Pirog, A.; Bedran, G.; Alfaro, J.; Dapic, I. Mass Spectrometry-Based Identification of MHC-Associated Peptides. Cancers 2020, 12, 535.
- Okada, M.; Shimizu, K.; Fujii, S.I. Identification of Neoantigens in Cancer Cells as Targets for Immunotherapy. Int. J. Mol. Sci. 2022, 23, 2594.
- Kotsias, F.; Cebrian, I.; Alloatti, A. Antigen processing and presentation. Int. Rev. Cell Mol. Biol. 2019, 348, 69–121.
- Freudenmann, L.K.; Marcu, A.; Stevanovic, S. Mapping the tumour human leukocyte antigen (HLA) ligandome by mass spectrometry. Immunology 2018, 154, 331–345.
- Peters, H.L.; Tripathi, S.C.; Kerros, C.; Katayama, H.; Garber, H.R.; St John, L.S.; Federico, L.; Meraz, I.M.; Roth, J.A.; Sepesi, B.; et al. Serine Proteases Enhance Immunogenic Antigen Presentation on Lung Cancer Cells. Cancer Immunol. Res. 2017, 5, 319–329.
- Zhang, X.; Qi, Y.; Zhang, Q.; Liu, W. Application of mass spectrometry-based MHC immunopeptidome profiling in neoantigen identification for tumor immunotherapy. Biomed. Pharm. 2019, 120, 109542.
- Santambrogio, L. Molecular Determinants Regulating the Plasticity of the MHC Class II Immunopeptidome. Front. Immunol. 2022, 13, 878271.
- Olsson, N.; Jiang, W.; Adler, L.N.; Mellins, E.D.; Elias, J.E. Tuning DO:DM Ratios Modulates MHC Class II Immunopeptidomes. Mol. Cell Proteom. 2022, 21, 100204.
- Alspach, E.; Lussier, D.M.; Miceli, A.P.; Kizhvatov, I.; DuPage, M.; Luoma, A.M.; Meng, W.; Lichti, C.F.; Esaulova, E.; Vomund, A.N.; et al. MHC-II neoantigens shape tumour immunity and response to immunotherapy. Nature 2019, 574, 696–701.
- Hunt, D.F.; Henderson, R.A.; Shabanowitz, J.; Sakaguchi, K.; Michel, H.; Sevilir, N.; Cox, A.L.; Appella, E.; Engelhard, V.H. Characterization of peptides bound to the class I MHC molecule HLA-A2.1 by mass spectrometry. Science 1992, 255, 1261–1263.
- Yewdell, J.W. MHC Class I Immunopeptidome: Past, Present & Future. Mol. Cell Proteom. 2022, 100230.
- Nielsen, M.; Ternette, N.; Barra, C. The interdependence of machine learning and LC-MS approaches for an unbiased understanding of the cellular immunopeptidome. Expert Rev. Proteom. 2022, 1–12.
- Leko, V.; Rosenberg, S.A. Identifying and Targeting Human Tumor Antigens for T Cell-Based Immunotherapy of Solid Tumors. Cancer Cell 2020, 38, 454–472.
- Liepe, J.; Sidney, J.; Lorenz, F.K.M.; Sette, A.; Mishto, M. Mapping the MHC Class I-Spliced Immunopeptidome of Cancer Cells. Cancer Immunol. Res. 2019, 7, 62–76.
- Mishto, M. Commentary: Are There Indeed Spliced Peptides in the Immunopeptidome? Mol. Cell Proteom. 2021, 20, 100158.
- Mishto, M. What We See, What We Do Not See, and What We Do Not Want to See in HLA Class I Immunopeptidomes. Proteomics 2020, 20, e2000112.
- Katayama, H.; Kobayashi, M.; Irajizad, E.; Sevillarno, A.; Patel, N.; Mao, X.; Rusling, L.; Vykoukal, J.; Cai, Y.; Hsiao, F.; et al. Protein citrullination as a source of cancer neoantigens. J. Immunother. Cancer 2021, 9.
- Wei, J.; Zanker, D.; Di Carluccio, A.R.; Smelkinson, M.G.; Takeda, K.; Seedhom, M.O.; Dersh, D.; Gibbs, J.S.; Yang, N.; Jadhav, A.; et al. Varied Role of Ubiquitylation in Generating MHC Class I Peptide Ligands. J. Immunol. 2017, 198, 3835–3845.
- Beresford, G.W.; Boss, J.M. CIITA coordinates multiple histone acetylation modifications at the HLA-DRA promoter. Nat. Immunol. 2001, 2, 652–657.
- McGinty, J.W.; Marre, M.L.; Bajzik, V.; Piganelli, J.D.; James, E.A. T cell epitopes and post-translationally modified epitopes in type 1 diabetes. Curr. Diab. Rep. 2015, 15, 90.
- Sidney, J.; Vela, J.L.; Friedrich, D.; Kolla, R.; von Herrath, M.; Wesley, J.D.; Sette, A. Low HLA binding of diabetes-associated CD8+ T-cell epitopes is increased by post translational modifications. BMC Immunol. 2018, 19, 12.
- Drazic, A.; Myklebust, L.M.; Ree, R.; Arnesen, T. The world of protein acetylation. Biochim. Biophys. Acta 2016, 1864, 1372–1401.
- Robert, D.; Schreiber, L.J.O.; Smyth, M.J. Cancer Immunoediting: Integrating Immunity’s Roles in Cancer Suppression and Promotion. Science 2011, 331, 1565–1570.
- Chong, C.; Coukos, G.; Bassani-Sternberg, M. Identification of tumor antigens with immunopeptidomics. Nat. Biotechnol. 2022, 40, 175–188.
- Sellars, M.C.; Wu, C.J.; Fritsch, E.F. Cancer vaccines: Building a bridge over troubled waters. Cell 2022, 185, 2770–2788.
- Gardner, A.; Ruffell, B. Dendritic Cells and Cancer Immunity. Trends Immunol. 2016, 37, 855–865.
- Stoitzner, P.; Romani, N.; Rademacher, C.; Probst, H.C.; Mahnke, K. Antigen targeting to dendritic cells: Still a place in future immunotherapy? Eur. J. Immunol. 2022, 1–16.
- Brentville, V.A.; Metheringham, R.L.; Gunn, B.; Symonds, P.; Daniels, I.; Gijon, M.; Cook, K.; Xue, W.; Durrant, L.G. Citrullinated Vimentin Presented on MHC-II in Tumor Cells Is a Target for CD4+ T-Cell-Mediated Antitumor Immunity. Cancer Res. 2016, 76, 548–560.
- Cook, K.; Daniels, I.; Symonds, P.; Pitt, T.; Gijon, M.; Xue, W.; Metheringham, R.; Durrant, L.; Brentville, V. Citrullinated alpha-enolase is an effective target for anti-cancer immunity. Oncoimmunology 2018, 7, e1390642.
- Brentville, V.A.; Vankemmelbeke, M.; Metheringham, R.L.; Durrant, L.G. Post-translational modifications such as citrullination are excellent targets for cancer therapy. Semin. Immunol. 2020, 47, 101393.
- Brentville, V.A.; Metheringham, R.L.; Daniels, I.; Atabani, S.; Symonds, P.; Cook, K.W.; Vankemmelbeke, M.; Choudhury, R.; Vaghela, P.; Gijon, M.; et al. Combination vaccine based on citrullinated vimentin and enolase peptides induces potent CD4-mediated anti-tumor responses. J. Immunother. Cancer 2020, 8, e000560.
- Symonds, P.; Marcu, A.; Cook, K.W.; Metheringham, R.L.; Durrant, L.G.; Brentville, V.A. Citrullinated Epitopes Identified on Tumour MHC Class II by Peptide Elution Stimulate Both Regulatory and Th1 Responses and Require Careful Selection for Optimal Anti-Tumour Responses. Front. Immunol. 2021, 12, 764462.
- Choudhury, R.H.; Symonds, P.; Paston, S.J.; Daniels, I.; Cook, K.W.; Gijon, M.; Metheringham, R.L.; Brentville, V.A.; Durrant, L.G. PAD-2-mediated citrullination of nucleophosmin provides an effective target for tumor immunotherapy. J. Immunother. Cancer 2022, 10, e003526.
- Zarling, A.L.; Obeng, R.C.; Desch, A.N.; Pinczewski, J.; Cummings, K.L.; Deacon, D.H.; Conaway, M.; Slingluff, C.L., Jr.; Engelhard, V.H. MHC-restricted phosphopeptides from insulin receptor substrate-2 and CDC25b offer broad-based immunotherapeutic agents for cancer. Cancer Res. 2014, 74, 6784–6795.
- Engelhard, V.H.; Obeng, R.C.; Cummings, K.L.; Petroni, G.R.; Ambakhutwala, A.L.; Chianese-Bullock, K.A.; Smith, K.T.; Lulu, A.; Varhegyi, N.; Smolkin, M.E.; et al. MHC-restricted phosphopeptide antigens: Preclinical validation and first-in-humans clinical trial in participants with high-risk melanoma. J. Immunother. Cancer 2020, 8, e000262.
- Merikhian, P.; Darvishi, B.; Jalili, N.; Esmailinejad, M.R.; Khatibi, A.S.; Kalbolandi, S.M.; Salehi, M.; Mosayebzadeh, M.; Barough, M.S.; Majidzadeh, A.K.; et al. Recombinant nanobody against MUC1 tandem repeats inhibits growth, invasion, metastasis, and vascularization of spontaneous mouse mammary tumors. Mol. Oncol. 2022, 16, 485–507.
- Palitzsch, B.; Gaidzik, N.; Stergiou, N.; Stahn, S.; Hartmann, S.; Gerlitzki, B.; Teusch, N.; Flemming, P.; Schmitt, E.; Kunz, H. A Synthetic Glycopeptide Vaccine for the Induction of a Monoclonal Antibody that Differentiates between Normal and Tumor Mammary Cells and Enables the Diagnosis of Human Pancreatic Cancer. Angew. Chem. Int. Ed. Engl. 2016, 55, 2894–2898.
- Scheid, E.; Major, P.; Bergeron, A.; Finn, O.J.; Salter, R.D.; Eady, R.; Yassine-Diab, B.; Favre, D.; Peretz, Y.; Landry, C.; et al. Tn-MUC1 DC Vaccination of Rhesus Macaques and a Phase I/II Trial in Patients with Nonmetastatic Castrate-Resistant Prostate Cancer. Cancer Immunol. Res. 2016, 4, 881–892.
- Glaffig, M.; Stergiou, N.; Schmitt, E.; Kunz, H. Immunogenicity of a Fully Synthetic MUC1 Glycopeptide Antitumor Vaccine Enhanced by Poly(I:C) as a TLR3-Activating Adjuvant. ChemMedChem 2017, 12, 722–727.
- Trabbic, K.R.; Whalen, K.; Abarca-Heideman, K.; Xia, L.; Temme, J.S.; Edmondson, E.F.; Gildersleeve, J.C.; Barchi, J.J., Jr. A Tumor-Selective Monoclonal Antibody from Immunization with a Tumor-Associated Mucin Glycopeptide. Sci. Rep. 2019, 9, 5662.
- Yarmarkovich, M.; Marshall, Q.F.; Warrington, J.M.; Premaratne, R.; Farrel, A.; Groff, D.; Li, W.; di Marco, M.; Runbeck, E.; Truong, H.; et al. Cross-HLA targeting of intracellular oncoproteins with peptide-centric CARs. Nature 2021, 599, 477–484.
- Zarling, A.L.; Polefrone, J.M.; Evans, A.M.; Mikesh, L.M.; Shabanowitz, J.; Lewis, S.T.; Engelhardt, V.H.; Hunt, D.F. Identification of class I MHC-associated phosphopeptides as targets for cancer immunotherapy. Proc. Natl. Acad. Sci. USA 2006, 103, 14889–14894.
- Cobbold, M.; De La Pena, H.; Norris, A.; Polefrone, J.M.; Qian, J.; English, A.M.; Cummings, K.L.; Penny, S.; Turner, J.E.; Cottine, J.; et al. MHC Class I-Associated Phosphopeptides Are the Targets of Memory-like Immunity in Leukemia. Sci. Transl. Med. 2013, 5, 203ra125.
- Penny, S.A.; Abelin, J.G.; Malaker, S.A.; Myers, P.T.; Saeed, A.Z.; Steadman, L.G.; Bai, D.L.; Ward, S.T.; Shabanowitz, J.; Hunt, D.F.; et al. Tumor Infiltrating Lymphocytes Target HLA-I Phosphopeptides Derived From Cancer Signaling in Colorectal Cancer. Front. Immunol. 2021, 12, 723566.
- Singhal, A.; Fohn, M.; Hakomori, S. Induction of alpha-N-acetylgalactosamine-O-serine/threonine (Tn) antigen-mediated cellular immune response for active immunotherapy in mice. Cancer Res. 1991, 51, 1406–1411.
- Laubreton, D.; Bay, S.; Sedlik, C.; Artaud, C.; Ganneau, C.; Deriaud, E.; Viel, S.; Puaux, A.L.; Amigorena, S.; Gerard, C.; et al. The fully synthetic MAG-Tn3 therapeutic vaccine containing the tetanus toxoid-derived TT830-844 universal epitope provides anti-tumor immunity. Cancer Immunol. Immunother. 2016, 65, 315–325.
- Vlad, A.M.; Finn, O.J. Glycoprotein tumor antigens for immunotherapy of breast cancer. Breast. Dis. 2004, 20, 73–79.
- Padler-Karavani, V. Glycan Microarray Reveal the Sweet Side of Cancer Vaccines. Cell Chem. Biol. 2016, 23, 1446–1447.
- Marchiori, M.F.; Bortot, L.O.; Carvalho, I.; Campo, V.L. Synthesis of MUC1-derived glycopeptide bearing a novel triazole STn analog. Carbohydr. Res. 2020, 498, 108155.
- Trabbic, K.R.; Kleski, K.A.; Barchi, J.J., Jr. A Stable Gold Nanoparticle-Based Vaccine for the Targeted Delivery of Tumor-Associated Glycopeptide Antigens. ACS Biol. Med. Chem. Au. 2021, 1, 31–43.
- Stergiou, N.; Urschbach, M.; Gabba, A.; Schmitt, E.; Kunz, H.; Besenius, P. The Development of Vaccines from Synthetic Tumor-Associated Mucin Glycopeptides and their Glycosylation-Dependent Immune Response. Chem. Rec. 2021, 21, 3313–3331.
- Asín, A.; García-Martín, F.; Busto, J.H.; Avenoza, A.; Peregrina, J.M.; Corzana, F. Structure-based Design of Anti-cancer Vaccines: The Significance of Antigen Presentation to Boost the Immune Response. Curr. Med. Chem. 2022, 29, 1258–1270.
- Toraskar, S.; Madhukar Chaudhary, P.; Kikkeri, R. The Shape of Nanostructures Encodes Immunomodulation of Carbohydrate Antigen and Vaccine Development. ACS Chem. Biol. 2022, 17, 1122–1130.
- Ferreira, J.A.; Relvas-Santos, M.; Peixoto, A.; Silva, A.M.N.; Lara Santos, L. Glycoproteogenomics: Setting the Course for Next-generation Cancer Neoantigen Discovery for Cancer Vaccines. Genom. Proteom. Bioinform. 2021, 19, 25–43.
