1. Coccidioides Spp.
1.1. Taxonomy
Coccidioides immitis and
C. posadasii have been classified in a number of different ways over the years. In the 1890s
Coccidioides was initially classified as a protozoan, and the growth of a mold from a lesion was thought to be a contaminant
[5,6][1][2]. In 1905 Ophuls published a paper describing the pathology of the disease in detail, growing the organism, determining that it was a mold, and identifying spherules in human tissue and arthroconidia in mycelial culture
[7][3]. In addition to these mammoth accomplishments, he inoculated guinea pigs and rabbits with the mold and showed they developed lesions identical to the human pathology. This clearly established that coccidioidomycosis was a fungal disease and made the era of “protozoa” misidentification a very brief one.
The subsequent classification of the fungus was another issue. Through the late twentieth century the primary pathogenic fungi (
Blastomyces spp.,
Coccidioides spp.,
Histoplasma spp., and
Paracoccidioidomyces spp.) were classified as Deuteromycotina or Fungi Imperfecti because, at that time, the sexual state of those human pathogens had not been identified and sexual morphology was necessary for classification in the Kingdom Fungi. The application of DNA variation to fungal taxonomy eliminated the need for sexual features in taxonomy, and among the first “asexual” fungi shown to belong to the fungal kingdom and the phylum Ascomycota were the primary pathogenic fungi, including
Coccidioides [8][4]. Shortly thereafter, molecular taxonomy was applied to many pathogenic fungi, all of which could then be integrated into the Kingdom Fungi
[9][5]. The order Onygenales was proposed and included all the dimorphic fungal species known at that time, which caused invasive diseases in humans as well as the dermatophytes
[10,11][6][7]. Follow-up studies of a broader group of Onygenalean fungi by these and other investigators
[8,11][4][7] showed that non-pathogens were interspersed with pathogens throughout the Onygenales and confirmed that
Uncinocarpus reesii was closely related to
Coccidioides spp.
In 2004,
Blastomyces spp.,
Histoplasma spp., and
Paracoccidioidomyces spp. were reclassified as Ajellomycetacae by DNA homology studies, but
Coccidioides spp. did not fall into that group
[12][8]. Instead, it was most closely related to a group of non-pathogenic organisms that fell between Ajellomycetacae and the Gymnoascaceae clade (which contains dermatophytes). Another study using slightly different techniques to determine DNA homology also found that Ajellomycetacae and Onygenaceae (containing
Coccidioides spp. and a variety of non-pathogenic species) were sister groups
[13][9].
Whiston and Taylor took a more inclusive approach
[14][10]. They reasoned that it is difficult to determine relatedness of
Coccidioides spp. to non-pathogens when, among them, only the
U. reesii genome had been sequenced. To address this, they determined the genome sequence of
Amauroascus niger, Amauroascus mutatus,
Chrysosporium queenslandicum, and
Byssoonygena creatinophila and predicted the transcript assemblies/gene models
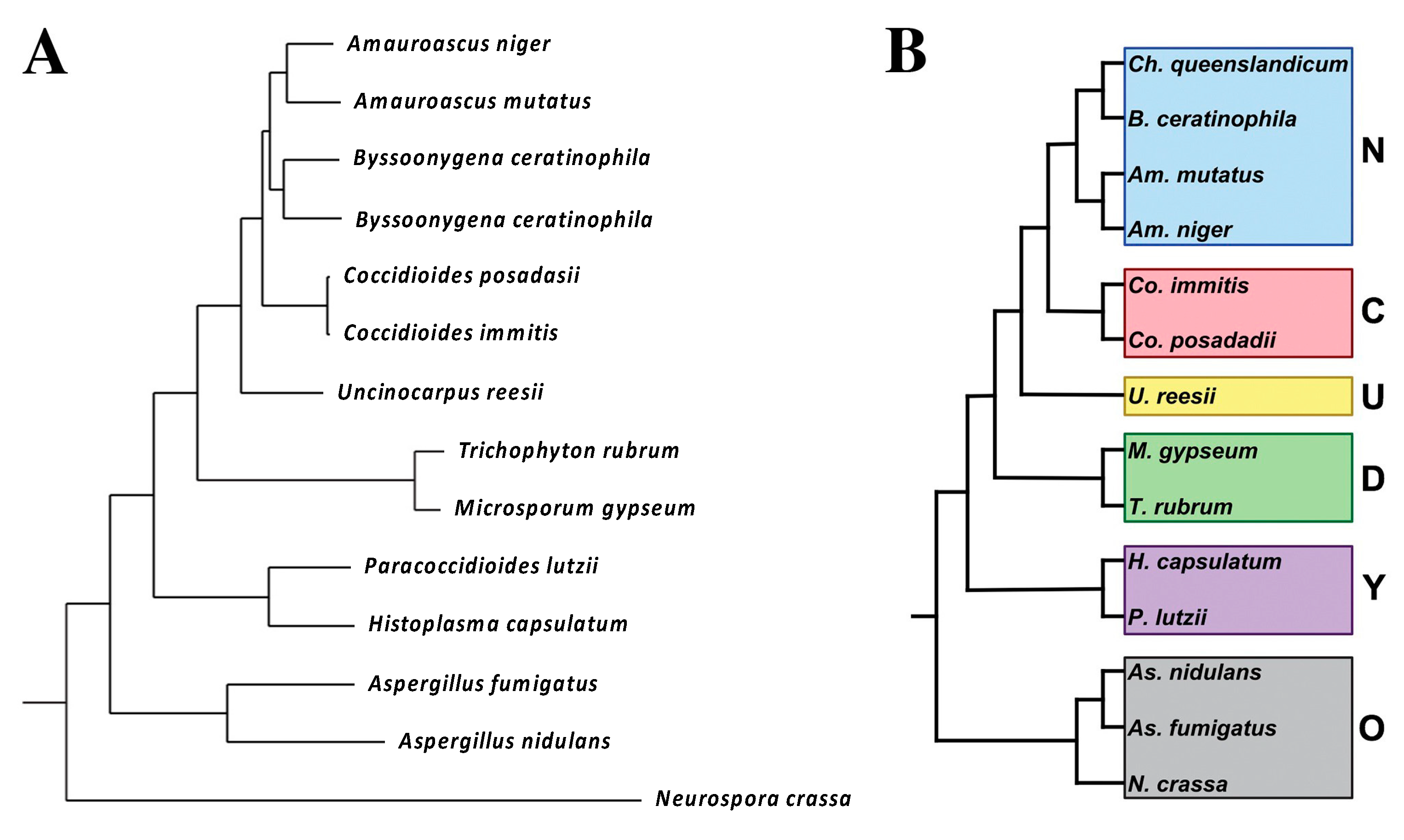
[14][10]. A phylogenetic tree was built by choosing 100 single-copy orthologs that were present in all the species, concatenating and aligning them. The phylogenetic tree is shown in
Figure 1.
Figure 1. Phylogenetic distance tree (Bayesian) with posterior probabilities based on 100 randomly selected single-copy orthologs (
A). Phylogenetic categories used for gene family expansion/contraction and ortholog group analysis (
B). N, newly sequenced genomes. C,
Coccidioides, U,
U.
reesii; D, dermatophytes; Y, yeast-forming dimorphic fungal pathogens; O, outgroups. The figure is from
[14][10].
The major conclusion
of th
is study erein is that at least five non-pathogenic organisms were closely related to
Coccidioides spp. A total of 791 genes were identified that were unique to
Coccidioides spp. These genes tend to be up-regulated in spherules (compared to mycelia)
[15[11][12],
16], suggesting that they might be important for differentiation into the pathogenic spherule phase.
More recently,
Coccidioides spp. has been placed within the family Onygenaceae, based on the DNA homology of the PRP8 gene
[17][13]. The only other pathogen in the Onygenaceae group was
Ophidiomyces ophiodiicola, which is a pathogen of snakes. The phylogenetic tree from their study is shown in
Figure 2. In this phylogeny, the family Onygenaceae is most closely related to the family Arthrodermataceae (containing the dermatomycetes), and both are slightly more distantly related to the family Ajellomycetaceae (containing the other systemic dimorphic pathogens
Histoplasma spp.,
Blastomyces spp., and
Paracoccidioides spp.); however, the statistical support for the basal branches determining these basal relationships is weak.
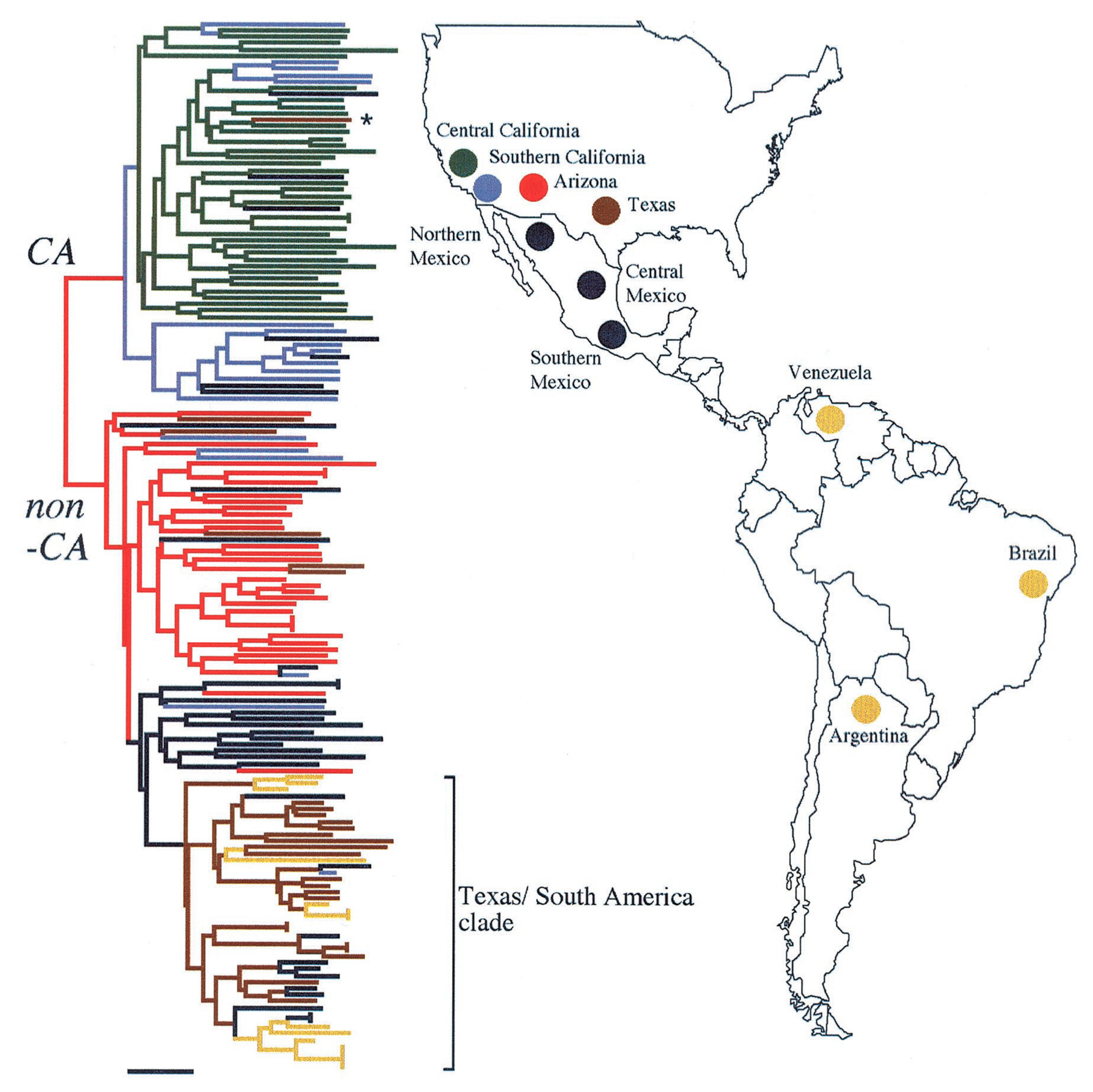
Figure 2. Geographic distribution of CA and non-CA strains. Biogeographic range expansion into South America by
C. immitis mirrors New World patterns of human migration. The colors refer to the genetic clusters on the left. * means: The asterisk marks an isolate from a patient who was diagnosed in Texas but had acquired the disease in California. The figure is from
[21][14].
What is the overall conclusion from all this effort? Although thermally dimorphic human pathogens cause similar diseases, they seem to be more genetically distinct (polyphyletic) than once imagined. It appears that the ability of Onygenales to cause invasive human infections has evolved at least twice, once in the Ajellomycetacae (
Blastomyces spp.,
Histoplasma spp., and
Paracoccidioides spp.) and once again in the Onygenaceae (
Coccidioides spp.). This also suggests that genetic programs underlying virulence in mammals may be significantly different in these two taxa and raises doubts about drawing conclusions based on homology between these human pathogens. The concept that thermal dimorphism and pathogenicity for humans has repeatedly evolved in
Coccidioides spp.,
Paracoccidioides spp.,
Histoplasma spp.,
Talaromyces marneffei, and
Sporothrix spp. has been previously discussed
[18][15].
C. Immitis and C. Posadasii
Until 1996,
C. immitis was thought to be the only species of
Coccidioides and it was thought to be an asexual organism. In that year Taylor and colleagues reported evidence of recombination and the division of the isolates into two geographic groups, one in California and the other in Arizona, Texas, and Mexico
[19,20][16][17]. These groups were initially referred to as CA and non-CA groups. Further studies of polymorphisms included many isolates from South America in the non-CA group, which proved to be more closely related to strains from Texas than strains from Arizona (
Figure 2)
[21][14].
Fisher and colleagues estimated that the two taxa had been reproductively isolated about 8 million years ago, although this estimate was later revised downward. Eventually these two taxa were named
C. immitis and
C. posadasii [22][18].
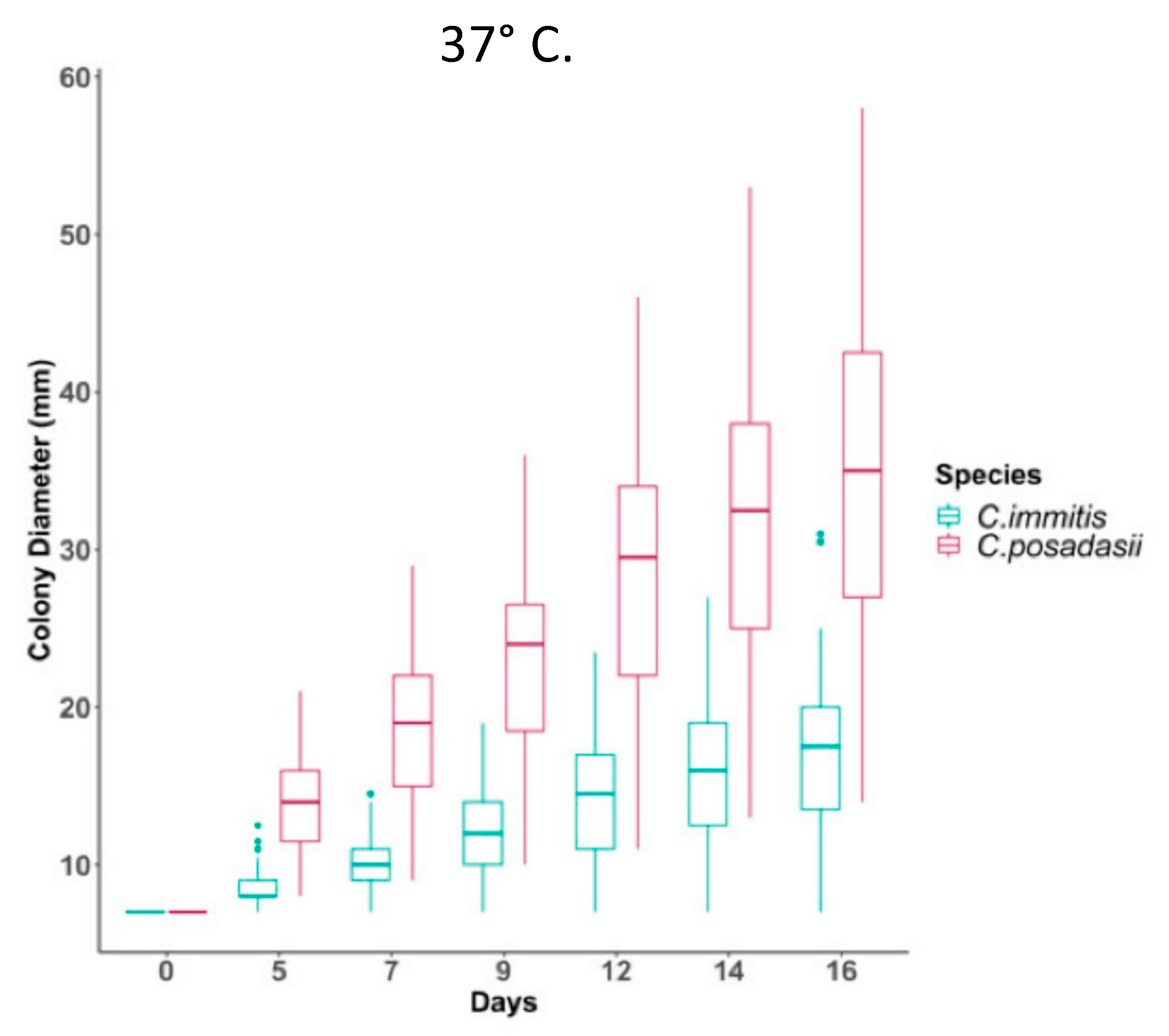
C. posadasii was able to grow as mycelia on high salt media more rapidly than
C. immitis. It has also recently been reported that
C. posadasii mycelia grow more rapidly at 37 °C than
C. immitis mycelia (
Figure 3)
[23][19].
Figure 3. Temperature impacts growth ability of
C. immitis isolates compared to
C. posadasii on yeast extract media. Radial growth measurements at 37 °C for 46
C. posadasii and 39
C. immitis isolates in triplicate. The figure is from
[23][19].
Differences in temperature effects on spherule growth have not been reported. Genome sequencing has confirmed the two species are distinct with some interspecific gene flow. As far as
weit is know
n, other major phenotypic differences between
C. immitis and
C. posadasii have not been reported, although that question has not been studied extensively. Certainly, serologic tests are cross-reactive and there is no evidence that protective immunity is species-specific. Since
Coccidioides spp. usually are not speciated in the clinical laboratory, it is currently difficult to assess any differences in the clinical human disease produced by the two species. Nevertheless, further studies will no doubt reveal more phenotypic differences between the two species. One observation that bears further study is the inability of experienced researchers to cultivate
C. posadasii from soil placed on growth media
[24][20], whereas direct cultivation of
C. immitis from soil has been routine for more than half a century
[25][21]. It is possible that
C. posadasii has evolved the ability to limit spore germination to mammalian lungs, thereby preventing germination in inhospitable environments, but no data have been published on this topic.
2. Geographic Distribution and Ecology
Coccidioides spp. are found in the desert and semi-arid soils of the Western hemisphere. The geographic limitations of these organisms were one of the first characteristics that were appreciated
[26,27][22][23]. The rate of positive skin test reactions in human beings has been used to estimate the presence of the organism in the soil. One of the early studies of coccidioidin skin test reactivity in military recruits found that the skin test positive rate was over 50% in some areas of California, Arizona, and Texas
[27][23]. The area of highest frequency in California was the San Joaquin Valley and in Arizona and Texas the highest rates were found in counties toward the south.
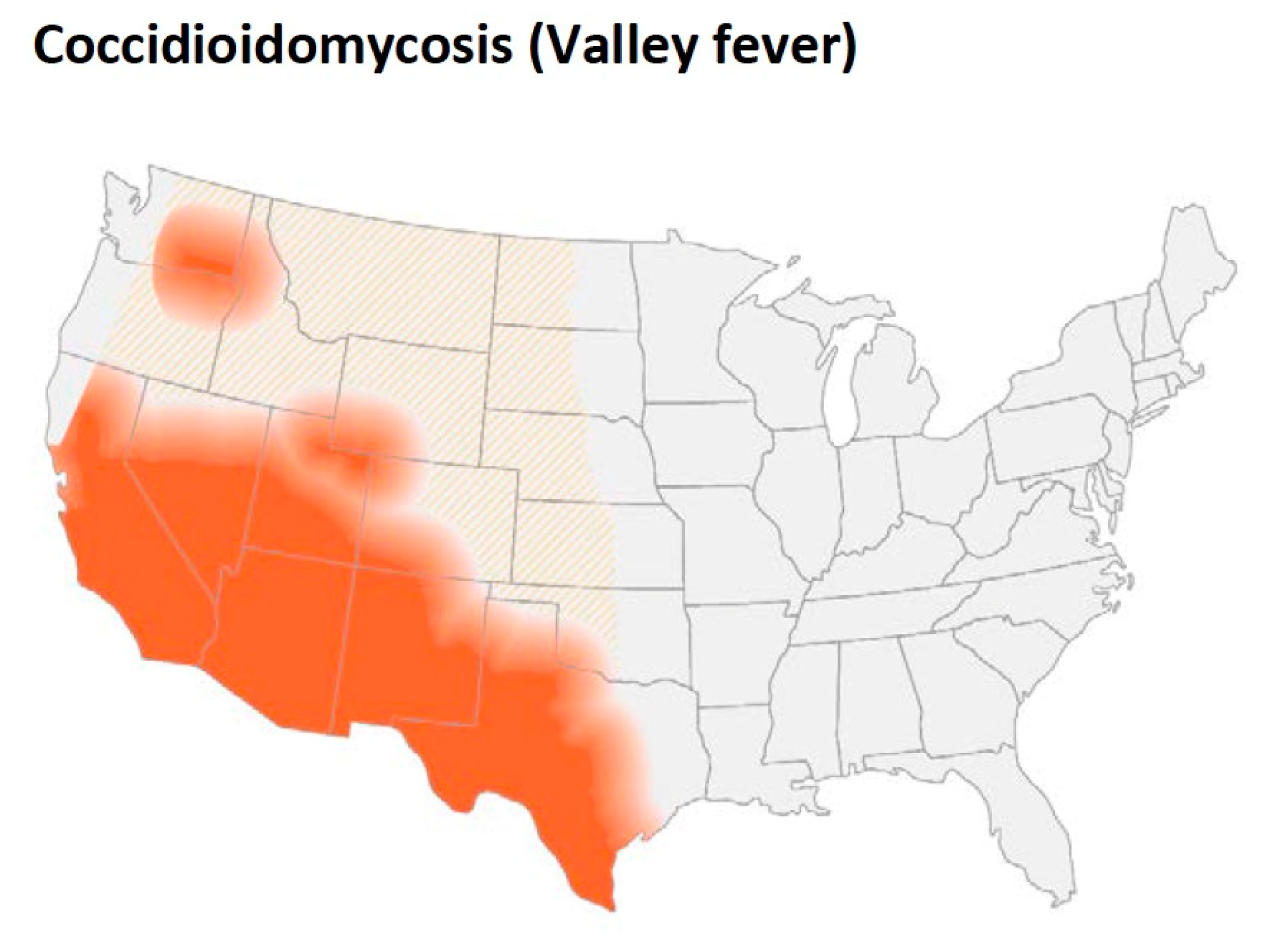
As
Figure 4 shows, in addition to California (especially the San Joaquin Valley), southern Arizona, southern New Mexico as well as west Texas are endemic regions. Smith pointed out that the organism was also endemic in areas in Argentina, Venezuela, and northern Mexico
[26][22]. He also reported more detailed geographic data about distribution of the disease in California. The incidence was maximal in the southern San Joaquin Valley, and some cases were seen in Monterey and San Luis Obispo counties. In fact, coccidioidomycosis is the most common fungal infection in stranded sea mammals in that area
[28][24]. In addition to these areas, small foci have been identified in north-eastern Utah and eastern Washington
[29,30,31,32][25][26][27][28].
Figure 4. Updated map from the CDC estimating the distribution of Coccidioides spp. in the United States. This figure is from the Centers for Disease Control and Prevention (https://www.cdc.gov/fungal/diseases/coccidioidomycosis/causes.html) (accessed on 10 August 2022).
The ecological factors associated with these geographic limitations have been investigated by many groups. It was first noted that the endemic region was in the Lower Sonoran Life Zone
[25][21]. This region corresponds to the hot deserts of the south-western United States and north-west Mexico (the Mojave, Sonoran, and Chihuahua deserts) where the soil is hot, dry, and alkaline. The vegetation includes the creosote bush (
Larrea tridentata) and the elevation of this region is between 100 ft to 3500–4000 ft. Total annual precipitation averages 10 inches or less. However, recovery of
Coccidioides spp. from the soil by standard fungal culture was generally very poor (0–15% of soil samples) and inconsistent from samples that were near each other. Recovery of
Coccidioides spp. seemed to be more common close to animal burrows
[33][29]. In addition, evaluation of desert rodents showed that some of them were infected both in Arizona
[34][30] and California
[25][21].
The incidence of clinical disease varies substantially from year to year, which is thought to reflect the amount of fungus in the soil. Weather is clearly an important factor; relatively wet winters followed by months with little or no rain are associated with high incidence of disease
[35,36,37][31][32][33]. A longitudinal study of a plot of soil also found that there were large differences in the recovery of
Coccidioides spp. from year to year
[38][34]. Recovery of
Coccidioides spp. was higher in years when high levels of sodium, calcium, chlorides, and sulfates were found in the topsoil.
Coccidioides spp. have not been reported from soil that is currently being farmed,
[39[35][36],
40], although farmworkers aren reported to be among the occupations at higher risk for coccidioidomycosis
[41][37].
A small number of clinical infections have been identified in the south-eastern region of Washington and
C. immitis has been detected in the soil by PCR and recovered by culture
[30][26]. A systematic study of
of soil in this area has been done
[42][38].
TheIt authoris found that the organism was present in several locations within an 46,000 m
2 area and persisted there for years. Soil with high levels of boron, calcium, magnesium, sodium, and silica was more likely to contain
C. immitis and, in laboratory experiments, serilized soil contained enough nutrients to support the growth of the organism. They reported no association of colonization with animal burrows, but reanalysis of the data while accounting for likelihood of detection showed that samples from animal burrows were more likely to be positive than those away from burrows
[43][39].
Another potential factor that is not usually considered is the presence of organisms in the soil that inhibit the growth of
Coccidioides spp. A study in Bakersfield California found that some bacteria isolated from local soil inhibited the growth of
C. immitis in vitro
[44][40]. The organisms with this activity were determined to be closely related to
Bacillus subtilis or
Streptomyces candidus. Presumably, these organisms in the soil could influence the amount of
C. immitis present and/or the ability to recover the fungus by culture. Other soil fungi probably play a role too.
Barker has recently reported that
C. posadasii is very difficult to recover from soil by culture in vitro
[24][20]. Even when
C. posadasii is spiked into sterile soil the colony recovery rate is only 0.2%. Therefore, in her hands, soil culture is not a useful tool for estimating the amount of
C.
posadasii. They also used a mouse inoculation technique to recover organisms. BALB/c mice were inoculated intraperitoneally with a saline extract of 5 g of soil and monitored for mortality or symptoms of infection and those that appeared to be infected were examined by histology and culture to prove that
Coccidioides spp. was the cause of their illness. Using this technique, 11/124 (9%) of the soil samples from the Tucson Arizona area contained
C. posadasii., so this is a much more sensitive technique than soil culture. Similar studies have not been done with
C. immitis, which can be cultured from soil
[25][21].
The use of PCR to detect fungal DNA in soil has been a major step forward since it is a very sensitive test for the detection of the organism. Several tests are available that use a variety of targets including the internal transcribed spacer 2 region, a transposon, and other genes
[45,46,47,48,49][41][42][43][44][45]. The sensitivity of the tests is largely dependent on the number of repeats of the gene target in the genome. In fact, the method of choice for detection of
Coccidioides spp. in environmental samples is a qPCR test based on a multi-copy transposon unique to the genus
[50][46]. Detection of
Coccidioides spp. in soil is routine, but detection from air is problematic
[51][47].
More recent ecological models have used much more sophisticated techniques
[52][48]. Several sites around Bakersfield California are known to contain
C. immitis DNA in the soil. A Landsat image of Kern County was used to evaluate the vegetation and soil temperature and a
Coccidioides-likelihood of detection score was calculated. In 25 sites there was a reasonably good correlation (75%) between a high likelihood score and detection of the organism in soil by PCR. The type of vegetation might be a proxy for the type of soil and climate conditions, but no detailed plant diversity data were presented to support that idea.
Another model used the maximum entropy algorithm to combine bio-climactic and geographic variables to estimate the likelihood of
Coccidioides spp. colonization
[53][49]. The predicted likelihood was compared to evidence of soil colonization by PCR and the correlation was excellent (AUC > 0.944). Predictions were also made for the geographic distribution of desert rodents:
Neotoma lepida (the desert woodrat) was the most closely associated rodent. The model predicts an increase in the geographic distribution of
Coccidioides spp. to the north and east as the environment gets warmer, and drier as is expected in the future.
To make predictions on a nation-wide scale, a model was developed using data from a variety of soil, temperature, and rainfall databases that spanned the entire U.S. and then building a fuzzy machine learning model
[54][50].
The auIt
hors found that the predicted is
uitability for growth of found that the predicted suitability for growth of Coccidioides spp. correlated fairly well with the areas of highest incidence of disease. Comparison to PCR studies of soil also showed reasonably good correlation. This model identified the most well-known endemic areas in the south-west, but it also predicted the area in eastern Washington State where there is an endemic focus, further validating the usefulness of this model.Desert animals are clearly infected with
Coccidioides spp. and evaluating evidence of infection in them is a very useful technique to evaluate the extent of endemicity
[24,33,55][20][29][51]. It has been proposed that persistence of the organism within granuloma in desert rodents and subsequent growth in the dead animal is a critical component of persistence in the environment
[55,56][51][52]. One modeling study has found that regions to be colonized with
Coccidioides spp. were also predicted to be colonized with the desert woodrat
[53][49]. Another animal that has been associated with infection in South America is the armadillo, where people who extracate this animal from burrows have become infected
[57][53] and
Coccidioides spp. has been cultured from a few animals
[58][54]. However, it is not clear how large a role animal infection plays in the persistence of the fungus in desert soils.
The small animal hypothesis is bolstered by the observation that gene families coding for enzymes that can digest animal proteins are expanded in
Coccidioides spp. genomes while genes for digestion of plants are lost (see
Section 6)
[59][55]. In addition, this hypothesis is attractive because it suggests a potential reason why the ability to differentiate into spherules and infect animals might be evolutionarily beneficial. The small animal hypothesis is certainly intriguing but does not preclude a role for growth outside animals. For example, there are reports that sterilized soil supports the growth of
Coccidioides spp.
[42,60][38][56] but hyphal growth in soil supported by rodent carcasses or other substrates could increase spore production and aid in dispersal as well as transmission to naïve hosts.
It is clear that the desert regions of the southwest U.S. and northern Mexico are highly endemic, but it is also clear that the organism is found in the soil of other semi-arid regions. In addition, the endemic region seems to be expanding. More studies of Coccidioides spp. in soil, infections in animals and epidemiologic studies are needed to better define the endemic areas and the risk for infection in those areas.
3. Culture, Detection and Morphology of Mycelia and Spherules
Coccidioides spp. grow well on many bacteriological media (including blood agar) and standard fungal culture media such as Sabouraud’s dextrose agar and colonies usually appear within several days. As with other dimorphic fungi, the fungus grows as a mold at room temperature to 35 °C at a pH of 5–8. The initial growth consists of hyphae without internal arthroconidia. If pigmentation occurs, it is usually on the underside of the colony
[61][57]. Arthroconidia usually develop within mycelia after one to two weeks but can take longer.
Coccidioides spp. arthroconidia are highly infectious so all manipulations of the organism or with suspected isolates should be done in a BSL 3 biosafety hood.
Coccidioides spp. can be identified in the appropriate clinical setting after the development of arthroconidia, but nucleic acid tests have been developed to identify the organism and, in some cases, to distinguish
C. immitis from
C. posadasii. The first molecular test was a DNA probe for identifying a colony as
Coccidioides spp. (
http://www.hologic.ca/products/clinical-diagnostics-and-blood-screening/assays-and-tests/accuprobe-culture-identification) (accessed 10 August 2022). There is one FDA-approved test for identification of
C. immitis and
C. posadasii DNA that distinguishes between the two species that has also been approved for direct testing of clinical specimens without culture (
https://genestatdiagnostics.com/coccidioidomycosis-valley-fever/) (accessed 10 August 2022). Several other tests are also available for detection of
Coccidioides DNA in clinical specimens
[45[41][42][43][44],
46,47,48], one of which also distinguishes between the two species
[49][45].
When the organism is grown in liquid media (usually in the research laboratory) the hyphal form is obtained
[62][58]. The most typical culture conditions are glucose yeast extract media at a temperature of 25–30 °C in the air with shaking. The maximal number of arthroconidia, develop after 4–6 weeks of culture when grown on solid medium.
3.1. Mycelia and Arthroconidia
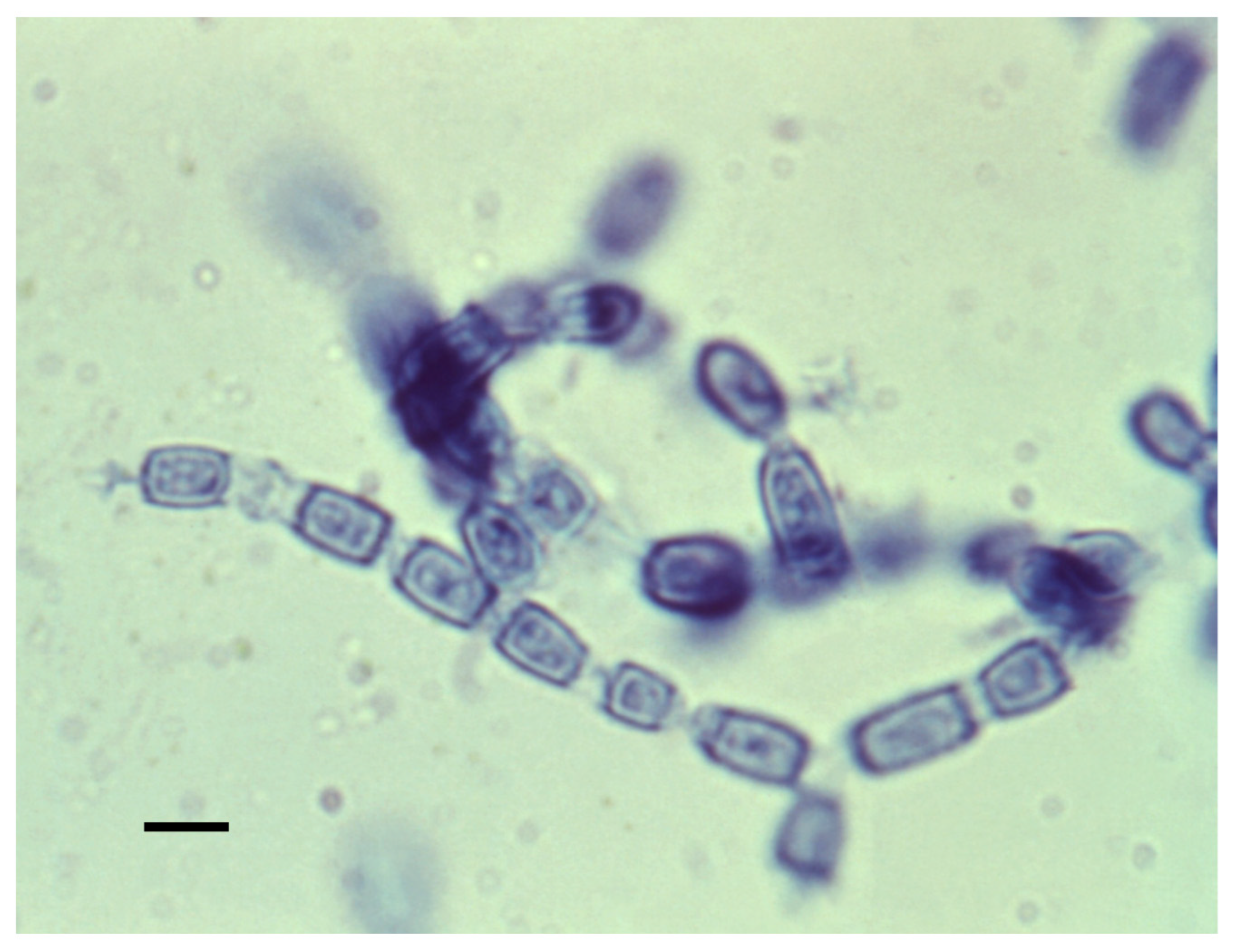
The mycelial form is shown in
Figure 5. Mycelia enlarge by apical growth followed by the development of septa. Once the septal spores close, every other cell degenerates, and the organism forms arthroconidia within a thin outer wall that is easily ruptured by the wind, resulting in their release. The arthroconidia contain two to five nuclei while the nuclei in the degenerating cell autolyze
[63][59]. The arthroconidia can either develop into mycelia (in the environment) or differentiate into spherules (in mammalian hosts).
Figure 5. Arthroconidia within mycelia. Every alternate cell has degenerated. The bar is 5 μM. The figure is from the Centers for Disease Control and Prevention. (https://phil.cdc.gov/Details.aspx?pid=15780) (accessed on 10 August 2022).
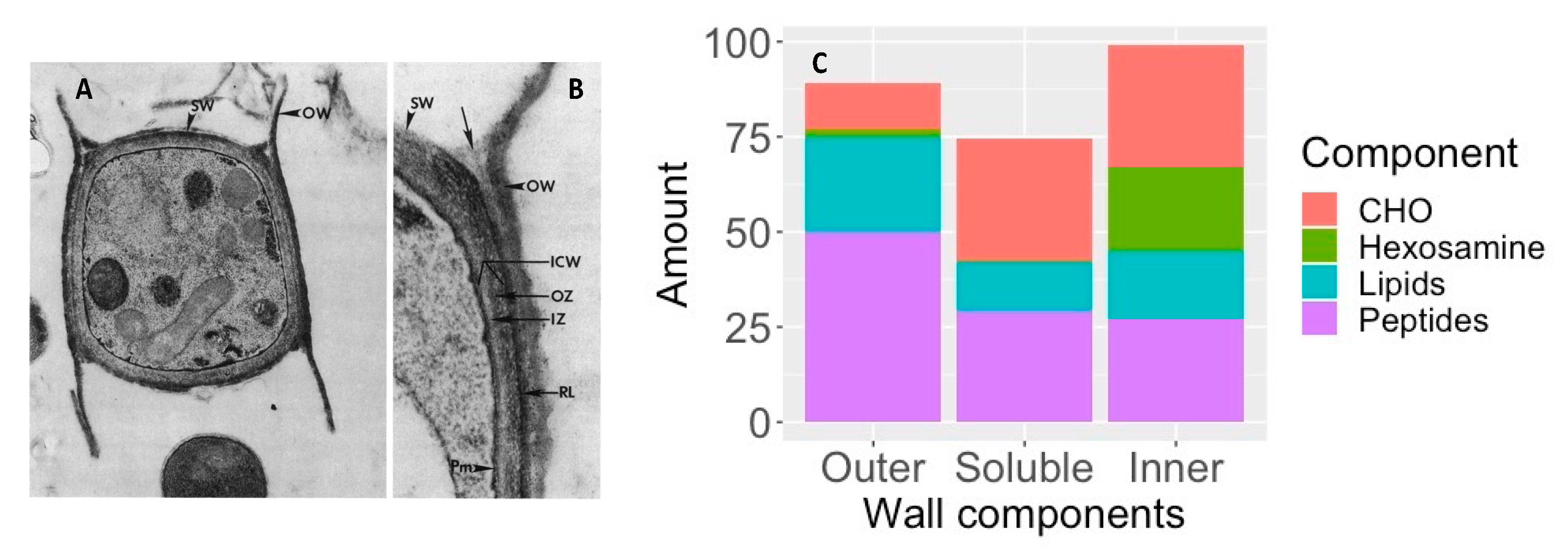
Arthroconidia have an inner and outer wall with an inner zone and a rodlet layer between the two as shown in
Figure 6A. The outer wall is a remnant of the outer sleeve of the mycelium. To determine the chemical composition, the outer wall and rodlet layer were sheared off together and separated from the soluble inner zone and the intact, viable organism
[64][60].
Figure 6C shows that the outer cell wall is rich in peptides and lipids, with relatively little carbohydrate, which probably contributes to its hydrophobic characteristics and its ability to become airborne and dispersed in the environment, including the airways of the mammalian host
[64][60]. The large amount of hexosamine in the inner wall is primarily contained in chitin. In addition, the unusual sugar 3-O-methylmannose was found to account for 12% of the carbohydrate in the inner wall. This sugar has not been found in any other fungi except
U. reesii [65,66][61][62]. Once arthroconidia are inhaled they begin to enlarge and their outer wall ruptures within 24 h. The soluble conidial wall fraction (SCWF) consists primarily of carbohydrates and peptides, with little lipid or hexosamine.
Figure 6. Components of arthroconidia cell wall. A and B: Thin sections of conidia of
C. immitis showing differentiation of wall layers. OW, outer wall layer; SW, septal walls of conidium; ICW, newly formed inner conidial wall composed of the outer zone (OZ) and more homogeneous inner zone (IZ); RL, rodlet layer; Pm, plasmalemma. Arrow locates soluble conidial wall fraction (SCWF) trapped between OW and RL which is released during the cell-shearing process. (
A), magnification ×12,000; (
B), magnification ×27,000. The figure is from
[64][60] and is reproduced with permission. (
C), composition of inner and outer cell wall and SCWF. The data for this figure is from
[64][60].
3.2. Spherules
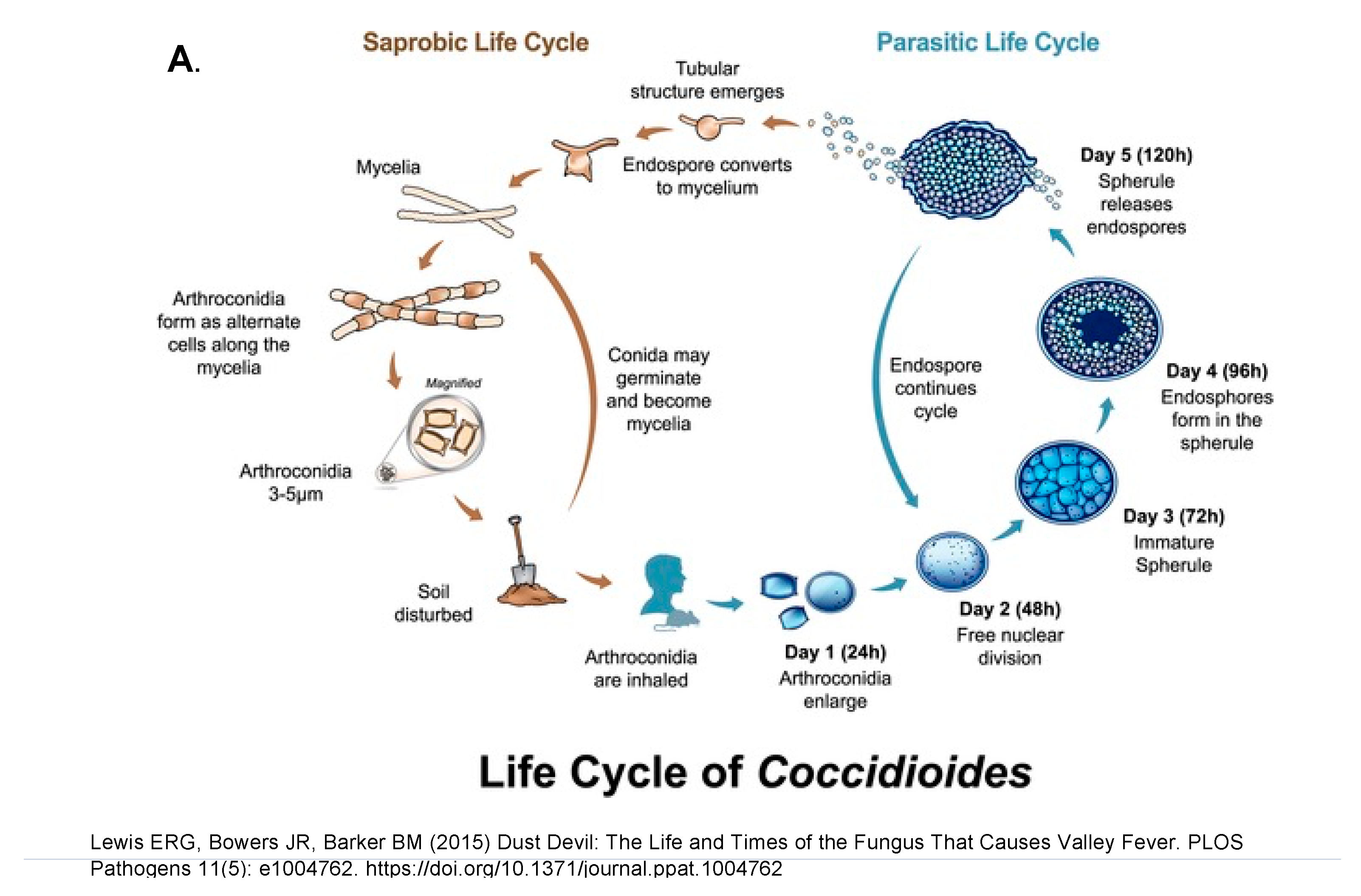
Differentiation into a spherule begins with the arthroconidium swelling into a round cell that then enlarges isotropically and divides internally to eventually form endospores. The round cell initially contains one nucleus, and the number of nuclei increases as internal division occurs leading to endospores with one nucleus each. A large spherule can contain hundreds of endospores. The spherule to endospore cycle begins when the spherule ruptures and releases endospores. Figure 7A represents the life cycle from soil to humans, while Figure 7B emphasizes the role of small mamals.
Figure 7. The saprobic (mycelia and arthroconidia) and parasitic (spherule and endospore) life cycle. The figure is from
[65][61]. (
A), mycelia growing in the soil and differentiating into spherules in animals. (
B), Endozoan-based life cycle of
Coccidioides species. Beginning at the asterisk (*), arthroconidia travel from the hyphae that produced them short distances among small mammals in burrows, or longer distances above ground, to infect other animals. The arthroconidia convert to spherules and are either controlled by the immune reaction or develop endospores, which disseminate to produce grave disease. The infected animal dies, either from disseminated coccidioidomycosis or from other causes and, in either case, living
Coccidioides present in the animal, now freed from the host immune system and living at lower temperatures, convert to hyphae. The hyphae grow through the dead animal and then produce abundant arthroconidia, which initiate a new cycle of life for the fungus. The figure is from
[55][51] and is reproduced with permission.
Spherule initials begin to divide by forming one cleavage plane, followed by another at 90° to the first, a third at 90° to the second, and so forth. The final stage of spherule development is the release of endospores by mature spherules. One group has reported that endospores are released in relatively large packets covered by fibrillar material
[67][63].
TheIt authoris hypothesized that the packets of endospores might be more difficult for white blood cells to phagocytose than single endospores. However, only a few studies of endospore phagocytosis have been reported. In general, endospores are a stage of the organism that has not been well studied.
Arthroconidia differentiate into spherules very quickly in vivo. Animal studies show that the conversion happens in the lung, in the muscle, or in the peritoneum within 24–48 h of inoculation. One clever way of obtaining spherules in vivo is to implant a chamber covered by a dialysis membrane subcutaneously into mice, wait until the chamber spontaneously fills with extravascular fluid and then inject arthroconidia
[68][64]. The chambers contain no mouse cells. Over the next ten days the organism grew as spherules (70%) and hyphae (30%). The addition of bronchial alveolar cells had no effect on the development of spherules. Removal of the chamber and culture at 37 °C in the air led to the development of hyphae but when cultured at 37 °C in 5% or 20% CO
2 a substantial number of spherules were obtained
[68][64]. Using buffers to obtain the same pH as was obtained in a CO
2 environment did not stimulate spherule growth.
TheIt authoris concluded that an elevated pCO
2 played a major role in spherule growth.
There are two general methods for growing spherules in vitro. One is the Converse medium, which is a synthetic medium of salts with glucose as the nutrient
[69][65]. Ammonium acetate is the primary salt, with NaCl, NaHCO
3 as well as potassium phosphate, calcium chloride and zinc sulfate present in lower concentrations. The pH is adjusted to 6.5. Usually the detergent Tamnol-N is added which is thought to enhance the release of endospores
[70][66]. A solid medium can be made by adding agar to the medium. The growth conditions are critical: a temperature of 35–40 °C and 5–15% CO
2 with shaking of liquid cultures is required. Mead and Barker have recently provided a detailed protocol for growing spherules
[62][58]. They recommend adding fresh medium if the spherules are maintained in culture for more than five days to facilitate endospore release. In some
C. immitis strains spherules develop within two days
[71][67] although most studies have used spherules after three to four days in culture
[15,72][11][68]. The pace of development of immature spherules to endospore release in vitro differs between experiments with values from four days to more than eight days which may be influenced by strain differences. After the release of endospores, the second generation of spherules becomes asynchronous quickly.
One group studied the effects of amino acids, vitamins and substituting other carbon sources on the conversion from arthroconidia to spherules
[73][69]. Phenylalanine, tyrosine, tryptophan, dihydroxyphenylalanine (DOPA) and pyrocatechol, singly or in combination, increased the rate of endosporulation. They argued that phenylalanine conversion to tyrosine, conversion of tyrosine to DOPA, followed by conversion of DOPA to melanin might be important for the formation of mature spherules.
Another study investigated the effect of human sex hormones on spherule growth since males and pregnant women in the third trimester are more likely to develop disseminated disease than the general population
[74][70]. Spherules, grown in Converse medium, were exposed to 17β-estradiol, 17α-estradiol, progesterone, or testosterone. All these hormones, except for 17α-estradiol, (which is an inactive stereoisomer) stimulated the growth of spherules at physiologic concentrations of hormones. Glucocorticoids did not stimulate spherule growth. 17β-estradiol also stimulated the growth of the organism in the mycelial phase. In a follow-up study, both a saturable high-affinity and low affinity-binding activity for progestin, estrogen, and androgen hormones were detected in spherule cytosol
[75][71].
RPMI is a medium that is designed for the growth of mammalian cells, but it also has been shown to support the growth of spherules. The initial study used RPMI with 10% fetal calf serum and 0.8 mg/mL N-Tamnol in 5% CO
2 at 35 °C with shaking
[76][72]. By 48 h, about 10% of organisms were in the spherule form. Hyphae were removed by filtration and spherules were collected by centrifugation. This process was repeated and after four passages essentially all the organisms were spherules. The N-Tamnol was important for rapid and complete conversion to spherules but once conversion occurred it could be eliminated. A recent study found that spherules grew larger and to a higher density in RPMI than in Converse medium and that growth in 1% O
2 (with CO
2) was preferable to air (with CO
2)
[72][68]. Other experiments have found that 10% fetal bovine serum or 0.1% Survanta in RPMI both support the growth of spherules
[77][73]. There is one study comparing spherules of several strains of
C. immitis and
C. posadasii by scanning electron microscopy
[72][68]. There appeared to be a difference in the surface of the spherules, with
C. posadasii Silveira having a more wrinkled surface than the other strains. Characterizing the differences between hyphae and spherules has been a major area of investigation
(see Section 8.1, Section 8.2 and Section 8.3).
Some methods for inducing the transformation of arthroconidia into spherules are well established but the detailed biological mechanisms of the differentiation are still a mystery. Some studies show that protein profiles of spherules induced by growth in Converse medium are different than spherules grown in RPMI (with different additives) or to spherules in vivo
[78][74]. These observations indicate the limitation of studies of spherules grown in vitro. In addition, there has been little investigation of the localization of proteins within the fungus during the transformation. Finally, there has been almost no investigation of endospore to spherule differentiation.