Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 3 by Dean Liu and Version 2 by Dean Liu.
Metastasis represents the most important cause of breast cancer-associated mortality. Even for early diagnosed stages, the risk of metastasis is significantly high and predicts a grim outcome for the patient. Nowadays, efforts are made for identifying blood-based biomarkers that could reliably distinguish patients with highly metastatic cancers in order to ensure a closer follow-up and a more personalized therapeutic method.
- breast cancer
- metastasis
- miRNA
- exosomes
1. Introduction
In 2020, GLOBOCAN indicated that breast cancer (BC) had become the leading cause of global cancer incidence worldwide, accounting for 11.7% of all cancers. Among women’s cancers, BC accounts for 25% of new cases and about 17% of deaths [1]. BC represents a highly heterogeneous disease with specific molecular, histological, and clinical features [2]. Gene expression profiling has enabled the molecular portraying of breast cancer which led to the PAM50 classification of breast tumors into five intrinsic subtypes: Luminal A and normal-like (characterized by ER+/PR+ and Ki67 low and good-to-intermediate prognosis), Luminal B (differentiated from Luminal A by high Ki67 and a decline in the patient’s prognosis), HER2+ (characterized by HER2 amplification and the lack of ER and PR), and TNBC (characterized by the lack of receptors ER−, PR−, and HER2−), with both the HER2+ and TNBC subtypes predicting a poor outcome [3]. While the clinical classification based on the expression of estrogen and progesterone receptors (ER, PR) and human epidermal growth factor receptor-2 (HER2) has utility in the selection of targeted therapies, the short-term patient responses and long-term survival remain difficult to predict [4]. In addition, each BC subtype possesses its own proliferation and metastasis capabilities, further hindering the patient’s outcome [5].
Despite new insights on molecular subtypes and the improvement of therapy management [6], about 90% of BC deaths are due to metastases [7][8]. BC metastasis represents a multistep evolutionary process involving a variety of cellular and molecular mechanisms. Shortly, this process is depicted as a series of sequential events by which: tumor cells leave their primary site, enter the lymphatic or blood stream, arrive by organotropism in specific secondary sites, and seed and colonize these sites by evading immune surveillance and acquiring resistance to therapeutic intervention [9]. Nevertheless, the molecular mechanism that underlies this process is very complex [10][11][12] and not yet fully elucidated.
Moreover the clinical–molecular landscape of BC metastasis has revealed two types of metastasis, referred to as de novo BC metastasis (dnBCM) and recurrent metastatic breast cancer (rMBC) [13]. DnBCM is identified in patients that present with stage IV disease at the time of diagnosis, while rMBC appears during tumor evolution after a while in patients initially diagnosed with locally advanced BC. The dnBCM has an incidence of about 6–10% of BC at the point of diagnosis, regardless of the improved screening programs [14]. A TCGA data analysis revealed significant clinical, pathological, and molecular differences between these types of BC metastasis, indicating that dnMBC patients are more responsive to the treatment, with better survival than rMBC patients [15][16]. Although difficult to predict, early diagnosed recurrences in BC patients have a 17–20% chance of improved survival [17].
Consequently, current data regarding the genomic drivers and phenotypic heterogeneity for each clinical BC subtype [18][19] were used to develop several recurrence gene expression-based predictors, including Oncotype Dx, MammaPrint, MapQuant D, Endopredict, the Breast Cancer Index (BCI), and PAM50-ROR [20]. Although gene expression tests have proven their usefulness for predicting the risk of recurrence of ER-positive BC patients, they have limitations in assessing the temporal heterogeneity of cancer and monitoring its evolution [21]. Therefore, identifying the phenotype alterations responsible for tumor evolution remains a challenging task.
A liquid biopsy represents the ideal method for investigating the dynamic BC phenotype of patients in order to identify their risk of relapse and the early detection of systemic dissemination. Previous data have pointed out the role of circulating tumor cells (CTCs), tumor-derived extracellular vesicles, circulating miRNAs, circulating tumor DNA (ctDNA), tumor-associated proteins, and tumor-educated blood platelets as promising and valuable biomarkers for a BC prognosis [22][23]. While CTCs and ctDNA are mostly associated with a liquid biopsy, they have some limitations in both sensitivity and specificity, especially for early cancer diagnostics [24].
A promising new class of BC biomarkers is arising in the form of miRNAs transported by tumor-delivered exosomes (miRNA-TDEx) [25][26]. MicroRNAs (miRNAs) are small, non-coding RNAs that regulate gene expression at the post-transcriptional level, being considered master modulators of the cancer phenotype. Therefore, miRNAs are involved in all the hallmarks of cancers, having oncogenic (oncomiR) or suppressor (tumor-suppressor miR) potential. The exosomes are small vesicles of approximately 100 nm, secreted by all cell types, with a role in local and systemic cell-to-cell communication. BC-derived exosomes are essential supporters of BC metastasis by modulating the phenotype of recipient cells and playing an important role in setting the pre-metastatic niche [27][28].
As aberrant miRNA expression is associated with the cancer phenotype, the identification of aberrantly expressed miRNAs in a liquid biopsy could be used to monitor the patient’s cancer status for the early detection of systemic recurrence. The discovery of new predictive miRNA-TDEx biomarkers in a liquid biopsy is essential for improving the management of breast cancer patients by periodical monitorization. Therefore, thires paper focuseearchers focus on identifying miRNA-TDEx with a proven role in BC metastasis prediction that could become useful in clinical practice. Moreover, considering the lack of standard methods for evaluating exosomal miRNA, researchers also discuss the challenges and the technical aspects underlying this issue. Therefore, researchers performed a thorough research of the literature presenting the potential role of exosomal miRNAs as biomarkers for metastasis prediction in breast cancer, up to May 2022. The PubMed database was queried using the following keywords: “miRNA exosomes breast cancer metastasis” and “miRNA extracellular vesicles breast cancer metastasis”. From the extensive list of retrieved papers, researchers focused on reviewing the research articles that evaluated aberrantly expressed tumor-derived exosomal miRNAs in metastatic breast cancer. The papers reporting miRNA evaluation from whole blood lysates or other types of vesicles, as well as small RNAs secreted by stromal or other cell types were excluded. The detailed search strategy is depicted in Figure 1.
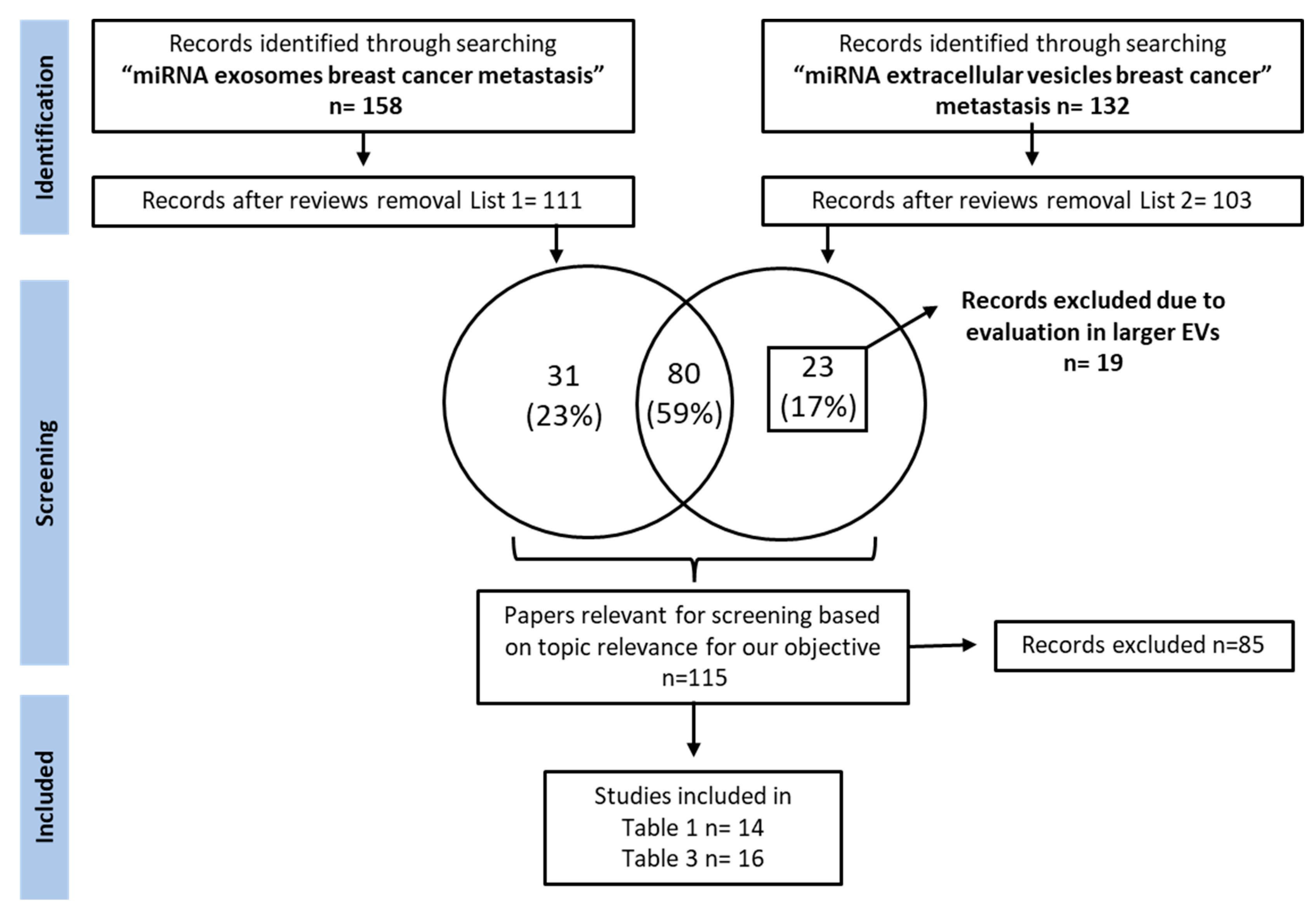
Figure 1. Study selection flow chart depicting the search strategy used for selecting papers.
A thorough research of PubMed literature was performed. Reviews were excluded from the retrieved results and papers were screened for relevant articles investigating the role of exosomal miRNAs as breast cancer metastasis biomarkers. MiRNAs of interest were grouped by their level of relevance to the clinic into two tables: one containing miRNAs with immediate applicability as liquid biopsy biomarkers (Table 1) and another one containing miRNAs validated in preclinical studies, with potential application to the clinical setting (Table 3).
2. Clinically Validated Exosomal miRNAs as Biomarkers for Breast Cancer Metastasis by Site
Breast cancer subtypes display different particularities with regard to therapy approaches, invasiveness, and survival rate. Moreover, each subtype develops a different metastatic pattern, with specific lesion rates in the secondary organs (Figure 2). Up to 70% [29] of metastases develop in the bone, which is the preferred niche for hormone receptor-positive (HR+) cancers [5]. Of all metastatic sites, bone metastasis tends to predict a better prognosis for the patient in terms of overall survival [30][31]. The liver represents the second most common metastatic site, affecting between 40–50% of all cases, and it is mostly associated with HR− and Her2+ breast cancers [32]. Patients with TNBC have the highest rate of lung and brain metastases [33][34]. Although brain metastasis accounts for the smallest percent of metastatic events in all types of breast cancers, 10–30% [33], more and more cases are being characterized with these particular lesions that unfortunately bare the grimmest overall survival rates [30][31]. Even though the metastatic disease is typically defined as the cancer spread to distant organs, the local progression of breast cancer to the lymph nodes represents a critical event that is indicative of a worse prognosis for the affected patients [35]. Even in the early stages, 10–26% of breast cancer patients diagnosed with T1 tumors present lymph node metastases which have been proven to be correlated with a higher risk of distant spread [36].
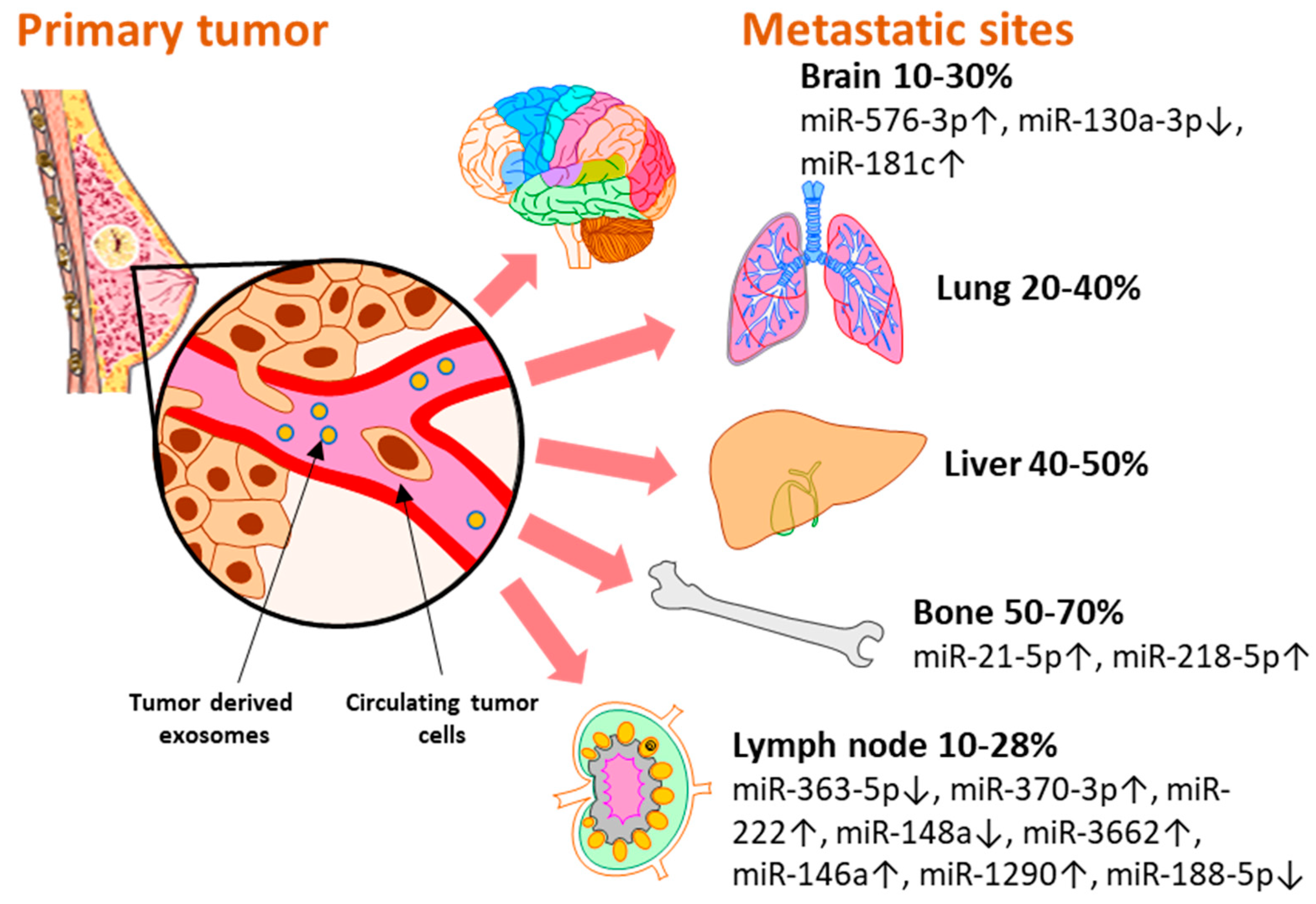
Figure 2. The distribution of miRNAs associated and validated with breast metastasis sites: brain (miR-576-3p, miR-130a-3p, miR-181c), bone (miR-21-5p, miR-218-5p), and lymph node (miR-363-5p, miR-370-3p, miR-222, miR148a, miR-3662, miR-146a, miR-1290, miR-188-5p). Breast cancer has several preferential metastasis sites, with different incidences of secondary lesions. Exosomal miRNAs released by tumor cells have been investigated as blood-based biomarkers for metastasis prediction with regard to the metastatic site. Several miRNAs reported in the literature have been validated in liquid biopsy samples of breast cancer patients and represent potential biomarkers with immediate applicability for the clinic.
MicroRNAs are key regulators of all the hallmarks of cancer [37][38] and their role as potential biomarkers for breast cancer progression and therapy response has also been reviewed elsewhere [39]. However, researchers believe it is noteworthy to highlight the importance and the potential use of exosome-encapsulated miRNAs in the clinical setting. Researchers present a set of exosomal miRNAs that were identified in liquid biopsy samples of breast cancer patients and were associated with metastatic events at different sites (Figure 2). As their biological functions were also validated in experimental settings, researcer consider that these miRNAs might represent potential biomarkers that could open new avenues for the early detection of breast cancer metastasis with minimal discomfort for the patient.
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249.
- Curtis, C.; Shah, S.P.; Chin, S.-F.; Turashvili, G.; Rueda, O.M.; Dunning, M.J.; Speed, D.; Lynch, A.G.; Samarajiwa, S.; Yuan, Y.; et al. The genomic and transcriptomic architecture of 2000 breast tumours reveals novel subgroups. Nature 2012, 486, 346–352.
- Dai, X.; Li, T.; Bai, Z.; Yang, Y.; Liu, X.; Zhan, J.; Shi, B. Breast cancer intrinsic subtype classification, clinical use and future trends. Am. J. Cancer Res. 2015, 5, 2929–2943.
- Rivenbark, A.G.; O’Connor, S.M.; Coleman, W.B. Molecular and cellular heterogeneity in breast cancer: Challenges for personalized medicine. Am. J. Pathol. 2013, 183, 1113–1124.
- Wu, Q.; Li, J.; Zhu, S.; Wu, J.; Chen, C.; Liu, Q.; Wei, W.; Zhang, Y.; Sun, S. Breast cancer subtypes predict the preferential site of distant metastases: A SEER based study. Oncotarget 2017, 8, 27990–27996.
- Gennari, A.; André, F.; Barrios, C.H.; Cortés, J.; de Azambuja, E.; DeMichele, A.; Dent, R.; Fenlon, D.; Gligorov, J.; Hurvitz, S.A. ESMO Clinical Practice Guideline for the diagnosis, staging and treatment of patients with metastatic breast cancer. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2021, 32, 1475–1495.
- Chaffer, C.L.; Weinberg, R.A. A perspective on cancer cell metastasis. Science 2011, 331, 1559–1564.
- Wang, L.; Zhang, S.; Wang, X. The Metabolic Mechanisms of Breast Cancer Metastasis. Front. Oncol. 2020, 10, 602416.
- Riggio, A.I.; Varley, K.E.; Welm, A.L. The lingering mysteries of metastatic recurrence in breast cancer. Br. J. Cancer 2021, 124, 13–26.
- Fares, J.; Fares, M.Y.; Khachfe, H.H.; Salhab, H.A.; Fares, Y. Molecular principles of metastasis: A hallmark of cancer revisited. Signal Transduct. Target. Ther. 2020, 5, 28.
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674.
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46.
- Seltzer, S.; Corrigan, M.; O’Reilly, S. The clinicomolecular landscape of de novo versus relapsed stage IV metastatic breast cancer. Exp. Mol. Pathol. 2020, 114, 104404.
- Heller, D.R.; Chiu, A.S.; Farrell, K.; Killelea, B.K.; Lannin, D.R. Why Has Breast Cancer Screening Failed to Decrease the Incidence of de Novo Stage IV Disease? Cancers 2019, 11, E500.
- Garrido-Castro, A.C.; Spurr, L.F.; Hughes, M.E.; Li, Y.Y.; Cherniack, A.D.; Kumari, P.; Lloyd, M.R.; Bychkovsky, B.; Barroso-Sousa, R.; Di Lascio, S. Genomic Characterization of de novo Metastatic Breast Cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2021, 27, 1105–1118.
- Lobbezoo, D.J.A.; van Kampen, R.J.W.; Voogd, A.C.; Dercksen, M.W.; van den Berkmortel, F.; Smilde, T.J.; Van De Wouw, A.J.; Peters, F.P.J.; Van Riel, J.M.G.H.; Peters, N.A.J.B. Prognosis of metastatic breast cancer: Are there differences between patients with de novo and recurrent metastatic breast cancer? Br. J. Cancer 2015, 112, 1445–1451.
- Lu, W.L.; Jansen, L.; Post, W.J.; Bonnema, J.; Van de Velde, J.C.; De Bock, G.H. Impact on survival of early detection of isolated breast recurrences after the primary treatment for breast cancer: A meta-analysis. Breast Cancer Res. Treat. 2009, 114, 403–412.
- Sotiriou, C.; Pusztai, L. Gene-expression signatures in breast cancer. N. Engl. J. Med. 2009, 360, 790–800.
- Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012, 490, 61–70.
- Prat, A.; Ellis, M.J.; Perou, C.M. Practical implications of gene-expression-based assays for breast oncologists. Nat. Rev. Clin. Oncol. 2011, 9, 48–57.
- Zeng, C.; Zhang, J. A narrative review of five multigenetic assays in breast cancer. Transl. Cancer Res. 2022, 11, 897–907.
- Heitzer, E.; Haque, I.S.; Roberts, C.E.S.; Speicher, M.R. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat. Rev. Genet. 2019, 20, 71–88.
- Venesio, T.; Siravegna, G.; Bardelli, A.; Sapino, A. Liquid Biopsies for Monitoring Temporal Genomic Heterogeneity in Breast and Colon Cancers. Pathobiol. J. Immunopathol. Mol. Cell. Biol. 2018, 85, 146–154.
- Fiala, C.; Diamandis, E.P. Circulating tumor DNA (ctDNA) is not a good proxy for liquid biopsies of tumor tissues for early detection. Clin. Chem. Lab. Med. 2020, 58, 1651–1653.
- Nakase, I.; Takatani-Nakase, T. Exosomes: Breast cancer-derived extracellular vesicles; recent key findings and technologies in disease progression, diagnostics, and cancer targeting. Drug Metab. Pharmacokinet. 2022, 42, 100435.
- Lakshmi, S.; Hughes, T.A.; Priya, S. Exosomes and exosomal RNAs in breast cancer: A status update. Eur. J. Cancer 2021, 144, 252–268.
- Tan, Y.; Luo, X.; Lv, W.; Hu, W.; Zhao, C.; Xiong, M.; Yi, Y.; Wang, D.; Wang, Y.; Wang, H. Tumor-derived exosomal components: The multifaceted roles and mechanisms in breast cancer metastasis. Cell Death Dis. 2021, 12, 547.
- Melo, S.A.; Sugimoto, H.; O’Connell, J.T.; Kato, N.; Villanueva, A.; Vidal, A.; Qiu, L.; Vitkin, E.; Perelman, L.T.; Melo, C.A. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell 2014, 26, 707–721.
- Chen, W.; Hoffmann, A.D.; Liu, H.; Liu, X. Organotropism: New insights into molecular mechanisms of breast cancer metastasis. NPJ Precis. Oncol. 2018, 2, 4.
- Wu, S.-G.; Li, H.; Tang, L.-Y.; Sun, J.-Y.; Zhang, W.-W.; Li, F.-Y.; Chen, Y.X.; He, Z.Y. The effect of distant metastases sites on survival in de novo stage-IV breast cancer: A SEER database analysis. Tumor Biol. J. Int. Soc. Oncodev. Biol. Med. 2017, 39, 1010428317705082.
- Kast, K.; Link, T.; Friedrich, K.; Petzold, A.; Niedostatek, A.; Schoffer, O.; Werner, C.; Klug, S.J.; Werner, A.; Gatzweiler, A. Impact of breast cancer subtypes and patterns of metastasis on outcome. Breast Cancer Res. Treat. 2015, 150, 621–629.
- Zhao, H.-Y.; Gong, Y.; Ye, F.-G.; Ling, H.; Hu, X. Incidence and prognostic factors of patients with synchronous liver metastases upon initial diagnosis of breast cancer: A population-based study. Cancer Manag. Res. 2018, 10, 5937–5950.
- Brosnan, E.M.; Anders, C.K. Understanding patterns of brain metastasis in breast cancer and designing rational therapeutic strategies. Ann. Transl. Med. 2018, 6, 163.
- Jin, L.; Han, B.; Siegel, E.; Cui, Y.; Giuliano, A.; Cui, X. Breast cancer lung metastasis: Molecular biology and therapeutic implications. Cancer Biol. Ther. 2018, 19, 858–868.
- Naxerova, K. Defining the role of lymph node metastasis in systemic breast cancer evolution. EBioMedicine 2020, 57, 102852.
- Zhao, Y.-X.; Liu, Y.-R.; Xie, S.; Jiang, Y.-Z.; Shao, Z.-M. A Nomogram Predicting Lymph Node Metastasis in T1 Breast Cancer based on the Surveillance, Epidemiology, and End Results Program. J. Cancer 2019, 10, 2443–2449.
- Abolghasemi, M.; Tehrani, S.S.; Yousefi, T.; Karimian, A.; Mahmoodpoor, A.; Ghamari, A.; Jadidi-Niaragh, F.; Yousefi, M.; Kafil, H.S.; Bastami, M. MicroRNAs in breast cancer: Roles, functions, and mechanism of actions. J. Cell. Physiol. 2020, 235, 5008–5029.
- Gomarasca, M.; Maroni, P.; Banfi, G.; Lombardi, G. microRNAs in the Antitumor Immune Response and in Bone Metastasis of Breast Cancer: From Biological Mechanisms to Therapeutics. Int. J. Mol. Sci. 2020, 21, E2805.
- Di Agostino, S.; Vahabi, M.; Turco, C.; Fontemaggi, G. Secreted Non-Coding RNAs: Functional Impact on the Tumor Microenvironment and Clinical Relevance in Triple-Negative Breast Cancer. Non-Coding RNA 2022, 8, 5.
More
