Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 3 by Camila Xu and Version 2 by Camila Xu.
Molecular imprinting (MI) is the most available and known method to produce artificial recognition sites, similar to antibodies, inside or at the surface of a polymeric material.
- molecularly imprinted layers
- surface polymerization
- electropolymerization
1. Introduction
Chemical and biochemical sensors are modern devices in analytical chemistry that simplify and miniaturize analytical determinations. Analytical methods based on modern sensor technology have solved many difficult analytical issues in research and society. Many scientific groups are working at the global level in the field and are reporting interesting results [1]. In general, conventional analytical methods have been extensively used due to their accuracy; nevertheless, most of these methods are expensive and require complex equipment, laboratory facilities, a high reagent consumption and well-trained personnel [2]. These drawbacks have prompted the research community to buildmore performant sensors for specific analysis of samples with complex matrices and very low concentrations, all at a lower reagent cost with inexpensive and easily-handled equipment, in order to perform in situ and on-site determinations [3].
Generally, a sensor is composed of three integrated parts (Figure 1), as follows: (i) a receptor for detecting the target analyte in a selective and sensitive manner, (ii) a physical transducer that converts the information obtained from the sensitive receptor into a measurable signal (usually an electric signal), and (iii) a suitable analytical device to process and show the significant signals formed by transducers and to calculate the results [4].
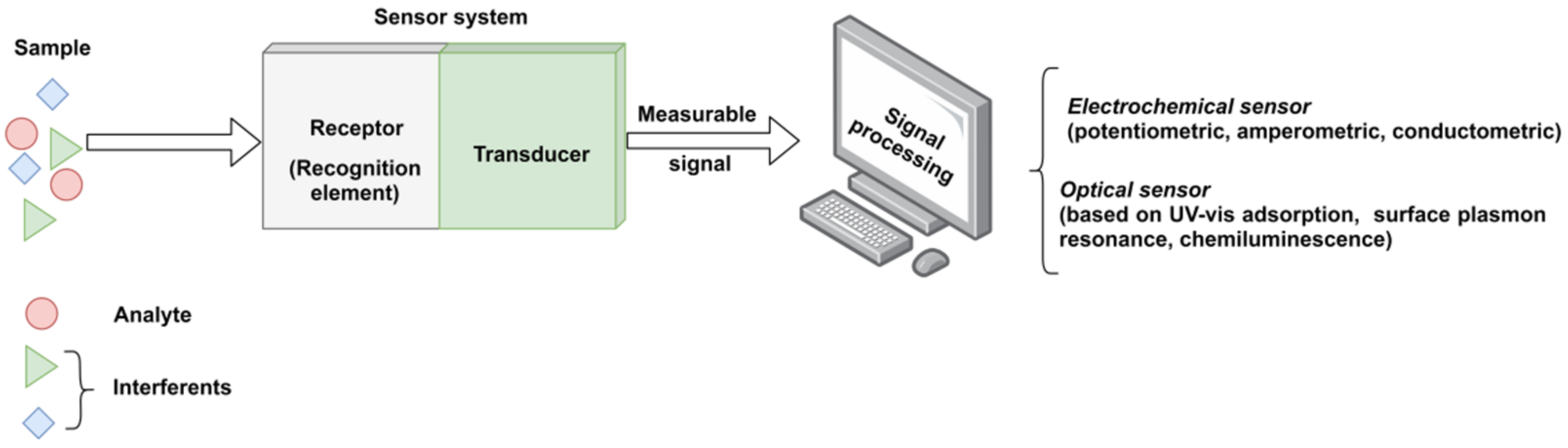
Figure 1. The functional scheme of a typical sensor system.
Biosensors are defined as analytical devices incorporating as a receptor biological material, such as enzymes, antibodies, nucleic acids, whole cells and tissues, a biologically derived material (i.e., engineered proteins, aptamers or recombinant antibodies) or a biomimetic material (i.e., molecularly imprinted polymers or combinatorial ligands). Depending on the incorporated sensitive material, the biosensors are classified as enzymatic, immune affinity recognition, DNA or whole-cell sensors; while considering the type of transducer, sensors can be classified as electrochemical, optical, acoustical, piezoelectrical, gravimetrical or thermal [1]. Each type of sensor class has other subclasses. For instance, the electrochemical sensors maybe amperometric (the majority), potentiometric, voltametric, etc. [5]. However, due to high production costs and the restricted operating conditions of these natural receptors, the development of artificial receptors with molecular recognition capacity, so-called synthetic receptors, have attracted a great interest as appropriate alternatives for the biological elements. Hence, among the existing techniques for the development of artificial receptors, high expectations are set out in the design of molecularly imprinted polymers (MIPs).
MIPs are polymeric materials that are designed and produced with built-in molecular recognition properties. As a result of this fundamental attribute, a growing interest has been observed in their development as inexpensive and robust materials with sensitive and selective molecular recognition. Some of the top current applications that include the use of MIPs are associated with catalysis, separation sciences and monitoring/diagnostic devices for chemicals, biochemicals and pharmaceuticals [6,7]. MIPs have proven to possess important advantages as an alternative sensing material for biosensors, including the ease of preparation, storage stability, low cost, repeated uses without loss of activity, high mechanical strength and resistance to heat and pressure as well as to harsh chemical environments [8,9]. In a typical approach, the MI process (Figure 2) allows the creation of specific molecular recognition sites by the polymerization of a functional monomer in the presence of target molecules (called template) and of a high concentration of crosslinker. Following the template removal, by specific extraction procedures, the specific recognition cavities are revealed [10,11,12].This approach endows MIPs with tremendous specific binding properties, as they possess cavities with complementary size, shape and electronic environment with the target molecule [13].
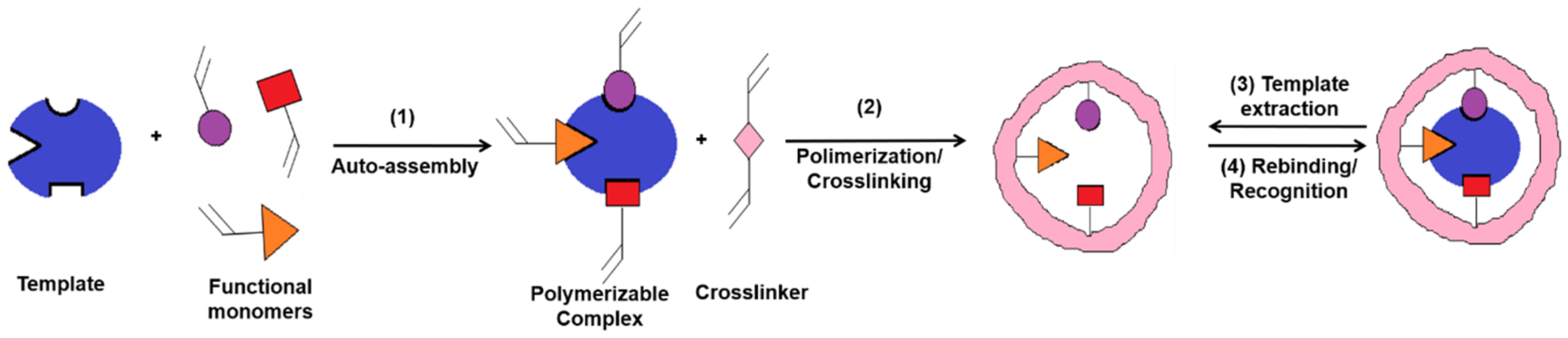
Figure 2. Molecular imprinting process (adapted from [14]).
In the classical imprinting process, a complex is formed between the template and functional monomer(s), by covalent, semi-covalent or non-covalent bonds, the non-covalent approach being preferred because of the simplicity in complex forming and the ease of the template extraction. In this case, polymerization takes place in the presence of an initiator and a porogen solvent, the latter having the role to create pores in the polymer matrix that facilitate the access of target molecules near the imprinted sites, during the rebinding assays [15]. A large amount of crosslinker is needed, as well, in order to stabilize the structure. Finally, the template is extracted from the crosslinked polymer and, thus, the imprinted cavities are created. This approach is also called the bulk method [16] because a solid block is first obtained, which is later on crushed for obtaining irregular-shaped particles. This method has little productivity because many of the recognition sites are destroyed during the crushing. This is the main reason why other methods were developed for enhancing the specificity and selectivity of MIPs, such as suspension [17], emulsion [18] or precipitation polymerization [19].
Molecular imprinting (MI) was applied initially for organic small molecule templates and ions [20], due to the molecular size, complexity, conformational flexibility and diffusion difficulties of large molecules [8]. Nevertheless, in the last 15 years, large molecules, such as proteins, were also successfully imprinted. For instance, by using the so-called epitope approach [21], only a characteristic part of the biomacromolecule is imprinted, and the following rebinding process is based on recognizing this part alone. Another clever approach developed from necessity allows the preparing of MIPs for labile, dangerous or very expensive targets, by performing the imprinting using a “dummy” template, meaning a safer or more available compound with a similar structure to that of the target analyte [22].
MIPs can also be used to design enzymatic and immunoaffinity sensors. In agreement with the classification of biosensors, MIP-based devices can work according to two different detection schemes: (1) affinity sensors (“plastic antibodies”) and (2) catalytic sensors (“plasticenzymes”) [2,23]. In this respect, MIP layers are usually deposited directly on the transducer surface of a sensor chip to produce the recognition unit [24,25]. As a result of this rather simple procedure, many researchers have found various methodologies for producing MIP layers as receptors for producing sensing devices, such as spin coating of a precursor solution to form a thin film, followed by insitu chemical polymerization [26]; electropolymerization of a pre-assembled complex of an electroactive functional monomer with a template [27,28]; dropcasting of a precursor polymer from solution [25,29,30]; dripping a composite solution containing a conducting material (e.g., graphene), MIP particles, and a binder (e.g., PVC) [31]; and, self-assembly of monolayers [32].
2. Molecular Imprinting by Surface Polymerization
In the surface imprinting process, the recognition sites are formed at the surface of a substrate [33]. Due to this fact, the recognition sites are more accessible and promote faster binding kinetics, compared to monolith MIPs for example. This means that template–polymer interactions are not governed by diffusion processes to the same extent as usually encountered in bulk imprinting [34]. Therefore, the technique is applicable especially for the imprinting of large biomolecules such as proteins [35,36], microorganisms and cells. Moreover, surface imprinting requires lower amounts of template molecules compared to the amounts used for other imprinting techniques because imprinting occurs right at the surface of the films [37].
In principle, the method consists of the preparation of a polymerization mixture, containing the functional monomer, the template, the initiator, the porogen solvent and the crosslinker, similar to the bulk method (see Figure 3). An amount of this mixture is cast on a solid surface, as for instance the sensor surface, and the formed thin film is polymerized thermically or by UV light curing; the latter being largely preferred. At the end, the template is extracted, thus generating the surface imprinted polymer [16].
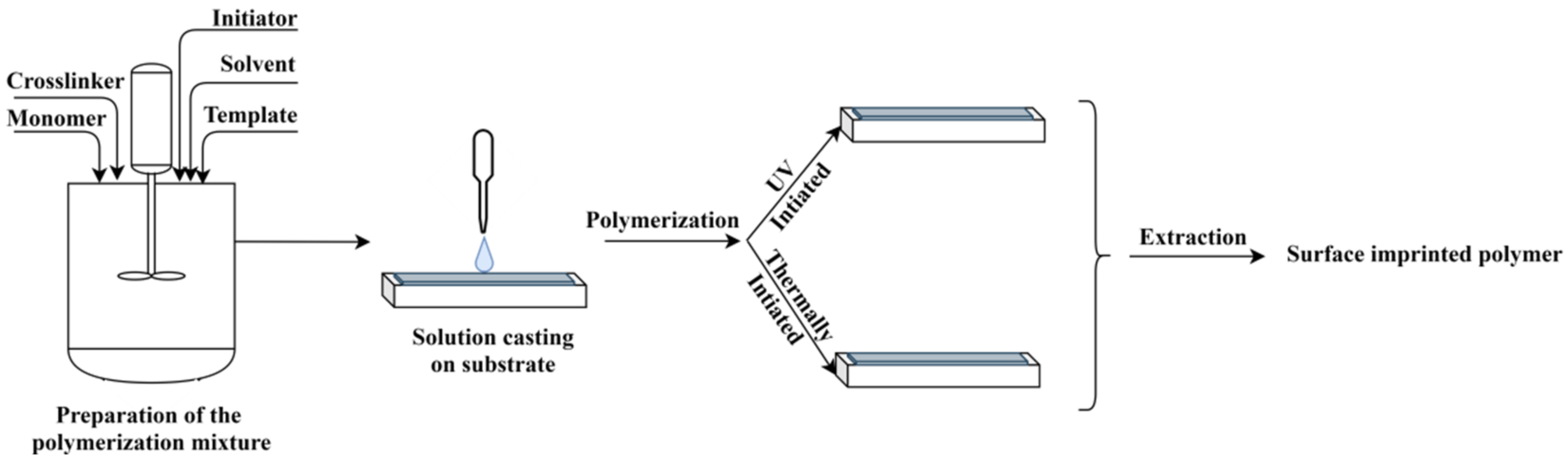

Figure 3. The principle of MIP layer preparation by surface polymerization.
Nonetheless, there are many variants of surface imprinting such as soft lithography, template immobilization, surface imprinting via grafting (pre-grafting polymerization and post-crosslinking/imprinting and grafting polymerization synchronized with crosslinking/imprinting [38]), emulsion and precipitation polymerization or epitope imprinting [16].
For example, Cennamo et al. [39], developed a biomimetic optical sensor based on MIP and surface plasmon resonance (SPR) transduction, in connection with tapered plastic optical fiber (POF) to detect selectively low molecular weight substances. The prepared SPR sensor was tested for l-nicotine ((−)-1-methyl-2-(3-pyridyl) pyrrolidine, MW = 162.24). In order to realize this SPR sensor, the plastic optical fiber, without protective jacket, was heated and stretched to yield a thinner part with a length of 10 mm, after which it was embedded in a resin block and a thin gold film was sputtered on its surface. According to the MIP classical procedure, the pre-polymerization mixture was prepared using l-nicotine as template, methacrylic acid (MAA) as functional monomer, divinylbenzene (DVB) as crosslinker, in a molar ratio of l-nicotine:MAA:DVB = 1:4:24. The developed device was able to be detected and discriminate between l- and d-nicotine using a small volume of sample, but with sensitivity strongly depending on the characteristics of the optical fiber [39]. Some examples of newsworthy MIP layersviathe grafting approach are listed in Table 1.
Table 1. MIP-based sensors obtained by surface polymerization.
| Synthesis Method | Receptor | Support | Analyte | Characterization Method(s) | LOD | Refs. |
|---|---|---|---|---|---|---|
| Spincoating | MIP film MIP nanoparticles |
Glass | Atrazine | RIfS 1 measurements. | >1.7 ppm | [40] |
| Precipitation polymerization/polymer casting | MIP layer | SERS substrate 2 | Enrofloxacin hydrochloride | Raman | 10−7 mol·L−1 | [41] |
| Casting | MIP membrane | Screen-printed gold electrode | Myoglobin | EIS 3 SWV 4 |
2.25 µg·mL−1 | [42] |
| Grafting polymerization synchronized with crosslinking/imprinting |
MIP film | GCE 5 | Olaquindox | CV 6, DPV 7, EIS | 7.5 nmol L−1 | [43] |
| Covalent imprinting/drop casting | MIP film | Au-TFME 8 | SARS-CoV-2 spike protein subunit S1 | CV, SWV | 4.8 pg·mL−1 | [44] |
| Dropcasting | MIP membrane | QCM 9 crystal chip | Human serum albumin | Langmuir, Freundlich, Langmuir–Freundlich isotherm |
0.026 μg mL−1 | [45] |
1 RIfS: reflectometric interference spectroscopy; 2 SERS: surface-enhanced raman scattering; silver nanoparticles modified by 3-methacryloxypropyltrimethoxysilane; 3 EIS: electrochemical Impedance spectroscopy; 4 SWV: square wave voltammetry; 5 glassy carbon electrode; 6 cyclic voltammetry; 7 differential pulse voltammetry; 8 MicruX™ gold-based thin-film metal electrodes; 9 QCM: quartz crystal microbalance.
Another optical sensor is described by Belmont et al. [40], in which case reflectometric interference spectroscopy (RIfS) was employed as a detection method, while the MIP films were prepared with the pesticide atrazine as the template molecule. In their study, the MIP films were deposited on glass transducers by two different methods: (i) spin-coating of pre-polymerization mixture containing polyvinyl acetate as a sacrificial polymeric porogen, followed by in situ surface polymerization of thin films, and (ii) auto-assembly of MIP nanoparticles with the aid of polyethylene imine (PEI) as an associative linear polymer. The results obtained upon assessment of atrazine solutions in toluene were reproducible for both types of films. However, the film prepared with auto-assembled MIP nanoparticles was more sensitive, tracking atrazine down to 1.7 ppm.
An example of the template immobilization approach is given in reference [42], where Moreira et al. developed a reusable sensor for Myo based on an MIP, prepared by assembling a polymer layer of carboxylated poly(vinyl chloride) (PVC COOH). This polymer was cast on the gold working area of a screen-printed electrode (Au-SPE), creating in this manner a novel disposable device relying on plastic antibodies.The MIP/Au-SPE sensor displayed a linear behavior by electrochemical impedance spectroscopy (EIS) and a limit of detection set-out at 2.25 μg/mL. The MIP/Au-SPE sensor also displayed good results in terms of selectivity. The error found for each interfering species were 11% for BSA, 7% for troponin T and2% for creatine kinase MB, respectively. Overall, the MIP modification over the Au-SPE was found suitable for producing an electrochemical sensor for screening Myo in biological fluids [43].
One epitope approach by surface polymerization for MIP selective layers is also illustrated by Ma and co-workers [44]. In the first step, the MIP was synthesized, using the epitope of human serum albumin as a template and afterwards, a coating method was applied to produce the quartz crystal microbalance sensor (EMIP-QCM). The MIP solution was prepared using zinc acrylateas a functional monomer, EGDMA as crosslinker and dimethylformamide (DMF) as porogen solvent. The gel precipitate was separated, washed and lyophilized. In order to obtain the sensor, a quartz crystal microbalance (QCM) crystal chip was used to drop-cast the MIP solution. The final MIP-QCM sensor displayed good selectivity and sensibility for human serum albumin, with a detection limit of 0.026 μg/mL. Furthermore, the sensor has also proven good accuracy and reproducibility when real samples were tested [45]. Further on, the study presented by Boysen and co-workers [46] is a typical example of a soft lithography approach and describes the design and synthesis of layer-by-layer MIPs via surface polymerization. The double-layered patterned MIP1/MIP2 was prepared on a silicon wafer. To enable chemical binding of MIP1 layer on the silicon, the surface of the wafer was first activated by sonication and exposed to UV-light, after which the surface was silanized with 3-(trimethoxysilyl)propyl methacrylate. The MIP pre-polymerization mixture was prepared by dissolving the template(N-dansyl-L-phenylalanine) and MAA with a crosslinker and a photo-initiator in a porogen solvent. This solution was spincoated onto the silanized silicon wafers and photo-polymerized. After template extraction, a 4-vinylpyridine-MIP thin film layer was deposited by photo-lithographic etching onto the first MIP film of PMAA, resulting in a grid-patterned surface in which two different MIPs, with pre-determined selectivity for N-dansyl-L-phenylalanine, alternated at a micron scale. Selectivity differences towards fluorescent template analogues were inspected using fluorescence microscopy [46].
MIP films can be prepared by surface polymerization also on organic support, such as multiwall carbon nanotubes (MWCNT) [47]. In this report, the used template was lysozyme (Lys) from egg white. The functional monomer was acrylamide (AAm), the crosslinker, methylene bis acrylamide (MBA) and the solvent, phosphate buffer (PBS). Besides using PBS for the protein protection, a redox initiation was employed consisting of ammonium persulfate (APS) and N,N,N′,N′-tetramethylethylenediamine (TEMED), allowing to perform the surface polymerization at room temperature. The selectivity assays showed thatthe Lys–MIP film registered higher capacity and affinity for Lys than for the other competitive proteins, such as cytochrome C (Cyt C), myoglobin (Myo), hemoglobin (Hb) and bovine serum albumin (BSA). The relative selectivity coefficients for Lys/Cyt C, Lys/Myo, Lys/Hb, and Lys/BSA were 1.30, 1.30, 3.12 and 2.82, respectively, while the adsorption capacity of the Lys–MIP film was 1.86 times higher than that of the non-imprinted polymer (NIP). Although the material was intended for selective separation of lysozyme, it may also be of interest for sensor application as a result of excellent electric conductivity of MWNT [47].
In recent decades, an extensive interest in the application of controlled radical polymerization methods (CRPs) for the imprinting of biomacromolecules has been observed [48]. Generally, MIPs are prepared by a conventional free radical polymerization mechanism, mainly due the fact that it is not disturbed by a large range of functional groups of the monomers and of the template, but also due to mild reaction conditions that can be employed. However, free radical polymerization has a major drawback due to the fact that chain propagation and termination reactions are hard to control, which makes the synthesis of surface imprinted polymer films with constant and targeted thickness difficult. Furthermore, free radical polymerization normally yields crosslinked polymer networks with heterogeneous structures, which might be responsible for the increased heterogeneity of binding sites, and thus, for the decreased affinity and selectivity [49]. On the other hand, the negligible chain termination in CRPs and their thermodynamically-controlled processes allow for more constant rates of the polymer chain growth, leading to more homogeneous polymer networks with narrower distributions of the chain length. This is the reason why several CRPs were developed so far for surface polymerization, in which case the most preferred methods refer to iniferter-induced radical polymerization [50,51], atom transfer radical polymerization (ATRP) [38,52] and reversible addition-fragmentation chain transfer (RAFT) polymerization [38,53]. Some of the successful procedures are provided next.
A selective surface plasmon resonance (SPR) sensor, based on surface polymerization, for the detection of Ochratoxin A (OTA) contamination in dried fig was developed by Akgönüllü et al. [54]. OTA is one of the most common mycotoxins that contaminate a wide range of agricultural products, which is why its assessment is very important for the monitoring of the food quality. The MIP layer was produced onto the SPR sensor chip by light-initiated polymerization of N-methacryloyl-L-phenylalanine (MAPA) and 2-hydroxyethyl methacrylate (HEMA) using OTA as a template. In a first step, the gold surface of the chip sensor was modified with allyl groups, by allyl mercaptan. This pre-treatment was performed because thiol-end will bind to the gold SPR chip, and the other end, allyl group, will interact with the polymer, insuring the good adherence of the MIP film on the chip. For the preparation of the MIP film, MAPA and OTA were mixed to obtain a pre-complex with the molar ratio of 1:3 for OTA:MAPA. The pre-polymerization complex was then mixed with HEMA, a crosslinker and a radical initiator in methanol. This reaction mix was dripped onto the SPR gold chip surface, distributed uniformly using a spin coater and, finally, polymerized by UV-light to produce the MIP film. The MIP–SPR chip was able to detect the OTA with high specificity (around 4.24 higher than that of the NIP), while the detection limit was close to 1 ng/mL and the response time was about 8 min [54].
Another interesting study on surface imprinting via the grafting (grafting from) method is provided by Heetal [55], which described the obtaining of a sensor for testosterone starting from porous silica by covalently binding azo-initiators and then photo-grafting. First, glycidoxy groups were immobilized on the surface of silica particles by the reaction of silanol groups with 3-glycidoxy-propyltrimethoxysilane (GPS), after which the glycidoxy groups were modified with an azo-initiator (4,4′-Azobis(4-cyanopentanoic acid)) to yield azo group-introduced silica particles. The following polymer grafting was carried out by photopolymerization of MAA as the functional monomer, ethylene glycol dimethacrylate (EGDMA) as the crosslinker, and testosterone as the template, on the surface of the azo-modified particle, which served as the initiator. The prepared particles with specific recognition ability for testosterone (imprinting factor of 1.52) were used for liquid chromatography [55]. Nevertheless, it is obvious that they can also be used for sensor development by nanoparticle auto-assembly [40].
In reference [56], a “grafting from” method is provided by Tarannum and Singh, who reported water-compatible surface imprinting of “baclofen” on silica surface. As a supporting matrix, a silica gel was used and the synthesis of the MIP for a skeletal muscle relaxant, namely baclofen (4-amino-3-p-chlorophenylbutyric acid) was carried out on the surfaces of silica gel. An imprinting network of sulfobetaine polyelectrolyte was prepared in aqueous medium only. This was grafted onto the silica gel matrix using 3-aminopropyltriethoxysilane (APTES) as a silane coupling agent and Michael addition reaction for further propagating the polymer grafting procedure. The rebinding studies showed that the MIP displayed good recognition for baclofen as compared to NIPs. Meanwhile, selectivity tests proved that MIP had a high affinity to baclofen in the presence of interferants (close structural analogues). Hence, a facile, specific, selective and water-compatible technique to detect baclofen in the presence of various interferants is provided. The prepared materials can be applied in HPLC as well as incapillary electrochromatography (CEC).
An interesting “dummy” imprinting approach by surface polymerization is described by Shahhoseini et al. [57]. In order to prepare sensitive layers for tricyclic antidepressants (TCAs) measurement in blood, the surface polymerization was performed on steel blades. In this respect, the pre-polymerization solution was obtained using a dummy template: benzyl(3-(10,11-dihydro-5H-dibenzo[b,f]azepin-5-yl)propyl)(methyl) carbamate, MAA and EGDMA and cast on the steel blades. Subsequently, the photo initiator was added and the layers were covered with a glass cover. The idea of using a dummy template, in this case, was derived from the necessity to prevent template bleeding. For trace analysis, such as the one targeted in this work, the risk of false positive results can also be due to incomplete template removal. Thus, to avoid such issues a pseudo-template was employed. The prepared MIPs were used as sensitive layers for liquid chromatography in which case the adsorption efficiencies for the MIPs were 3 to 5 times better than for the non-imprinted analogous, confirming that the use of the pseudo-template led to improved performance in this case.
The work of Hudson et al. [58] provides some insights regarding the surface molecular imprinting polymerization for obtaining a fluorescence sensor for the thermal and optical detection of nafcillin. The production of MIP films and particles was realized by integrating a fluorescent moiety that serves both as an element for template interaction and signaling. Fluorescein methacrylate (FluMa) was synthesized and introduced during the molecular imprinting process, first as the sole monomer and afterwards in an equimolecular mixture with MAA. The thin MIP films were deposited onto functionalized glass slides (to serve as electrodes) and the following UV-light initiated polymerization was performed in the presence of template species, nafcillin sodium salt.Although the specificity of films was not as high as that registered for the particles, the MIPs with FluMa and MAA were by far more performant than the ones with FluMa alone. Hence, the results are promising for developing a portable sensor for antibiotics [58].
3. Molecular Imprinting by Electropolymerization
Electrochemical polymerization is a method used to synthesize conductive polymers that are widely used for the development of biosensors and chemical sensors. This technique involves the deposition of a polymer layer on an electrode surface [59]. The electropolymerization can be performed by two methods, i.e., oxidation or reduction. Of the two methods, oxidation is the most commonly used for the obtaining conducting polymers. Anodic electropolymerization involves the monomer oxidation, thus obtaining cationic radicals, that lead to the formation of the polymer on the electrode surface [60,61]. Pyrrole, aniline and thiophene are the most important classes of conductive monomers, due to their low oxidation potential. Thus, by electropolymerization, polypyrrole, polyaniline and polythiophene are obtained. These polymers find their use in a wide range of domains because of certain advantages, referring to price, stability and synthesis complexity [62,63].
The electrochemical polymerization can be carried out using a three-electrode system (working electrode, counter electrode and reference electrode) or screen-printed electrodes (“3 in 1 electrode”). In a typical procedure, the following components are required: electrode system, solvent, supporting electrolyte, monomer(s) and template molecule (in case of MIP synthesis) [64]. The polymer layer deposition takes place on the working electrode (WE) surface (given schematically in Figure 4). The most common electrodes are made of gold, carbon and platinum, but other options are also available such as silver or indium tin oxide. Since, the properties of the polymer film are influenced by the electrode material, electrode surface, supporting electrolyte, electropolymerization technique, solvent and monomer, the overall procedure usually requires an optimization [65].
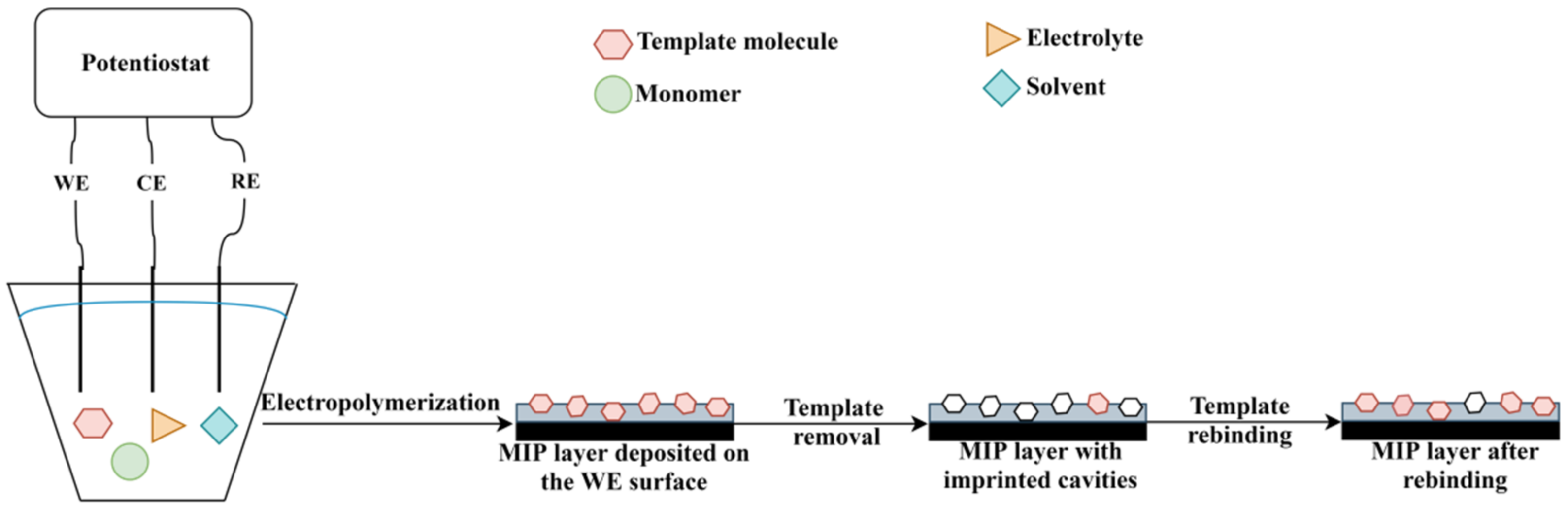

Figure 4.
The principle of MIP deposition by electropolymerization.
The electropolymerization can be performed using different techniques, including potentiodynamic (cyclic voltammetry), potentiostatic (constant potential) and galvanostatic (constant current) [66]. The potentiodynamic technique involves the formation of the polymer via cyclic voltammetry (CV). The polymer is formed upon applying a potential, which is changing between the oxidative and reductive state, resulting in a doped and undoped polymer [67]. On the other hand, potentiostatic polymerization is performed at a constant potential. In case of oxidative polymerization, the applied potential is positive, resulting in a doped polymer. In addition, the potential should be high enough to oxidize the monomer in order to initiate the polymerization process, but at the same time, it should be low enough to avoid secondary reactions [61]. Last but not least, the galvanostatic polymerization is carried out at a constant current, thus resulting in a doped polymer. This method has important advantages, such as simplicity, and most importantly, the thickness of the polymer depends on the electropolymerization time [68].
In recent decades, MIPs synthesized by electropolymerization have been widely used in the development of electrochemical sensors. An electrochemical sensor is a device consisting of a recognition element (in this case, MIP selective layer) and the electrochemical transducer. The principle of an electrochemical sensor relies on the interaction of the receptor with the analyte, which is transformed into an analytical signal (Figure 4). The main types of electrochemical sensors include conductometric, potentiometric, impedimetric and amperometric sensors [69,70]. Due to the improved selectivity, sensitivity and stability, limit of detection and low cost that MIPs provide, such MIP-based sensors are widely reported nowadays [71]. According to the recent literature, MIP-based electrochemical sensors are used in various applications in areas of human health, environmental pollution and pharmaceutical domain. The most significant results with MIP-based electrodes prepared by electropolymerization are provided in Table 2.
Table 2. MIP-based sensors obtained by electropolymerization.
MIP-based sensors obtained by electropolymerization.
| Synthesis Method | Receptor | Support | Analyte | Electrochemical Characterization Method(s) | LOD | Refs. |
|---|---|---|---|---|---|---|
| CV 1 | MIP film | Au electrode 2 | Acetaminophen | CV, EIS 3 and SWV 4 | 2.3 ×10−9 M | [72] |
| CV | MIP film | GCE 5 | Dimetridazole | DPV 6 | 10−10 M | [73] |
| Potentiostatic conditions | MIP film | Au electrode | Erythromycin | DPV | 1 × 10−10 M | [74] |
| CV | MIP film | Au electrode | Tetracycline | LSV 7 | 2.2 × 10−16 M | [75] |
| CV | MIP film | GCE | Sunset yellow | CV | 5 × 10−9 M | [76] |
| CV | MIP film | GCE | Tetra-bromo-bisphenol A | DPV | 2.7 × 10−10 M | [77] |
| CV | MIP film | Modified ITO 8 electrode | Resveratrol | CV and EIS | 7.1 × 10−12 M | [78] |
| CV | MIP film | Au electrode | Atrazine | CV and DPV | 1 × 10−9 M | [79] |
| CV | MIP film | Au electrode | Sodium lauryl sulfate | CV, DPV and EIS | 1.8 × 10−10 g/L | [80] |
| CV | MIP film | SPCE 9 | Diclofenac | DPV | 2 × 10−7 M | [81] |
| CV | MIP film | GCE | Epinephrine | CV and DPV | 7.6 × 10−8 M | [82] |
| CV | MIP film | Modified SPCE | Naloxone | DPV | 2 × 10−7 M | [83] |
| CV | MIP film | Modified SPCE | Naloxone | DPV | 1.6 × 10−7 M | [84] |
| CV | MIP film | Screen-printed gold electrode | Methylone | SWV | 1.1 × 10−6 M | [85] |
| CV | MIP film | Cr/Au film | Melamine | Resonance Wavelength Modulation | 5.1 × 10−12 M | [86] |
| CV | MIP film | GCE | Melamine | CV and SWV | 4.47 × 10−10 M | [87] |
| CV | MIP film | Modified GCE | Asulam | CV, DPV and EIS | 1.7 × 10−13 M | [88] |
| CV | MIP film | ITO electrode | Luteolin | DPV | 2.4 × 10−8 M | [89] |
| CV | MIP film | Modified electrode based on graphene and HAuCl4 | 4-nonylphenol | CV and DPV | 0.01 ng·mL−1 | [90] |
| CV | MIP film | GCE | Anthracene | SWV | 1.2 × 10−8 M | [91] |
| CV | MIP film | SPCE | Cocaine | SWV and EIS | 2.9 × 10−9 M | [92] |
| CV | MIP film | GCE | Entacapone | EIS and DPV | 5 × 10−8 M | [93] |
1 CV: cyclic voltammetry; 2 Au electrode: gold electrode; 3 EIS: electrochemical impedance spectroscopy; 4 SWV: square wave voltammetry; 5 GCE: glassy carbon electrode; 6 DPV: differential pulse voltammetry; 7 LSV: linear sweep voltammetry; 8 ITO: electrode indium tin oxide; 9 SPCE: screen-printed carbon electrode.
For instance, Menon and her co-workers [72] prepared an MIP-based electrochemical sensor for acetaminophen detection. Acetaminophen is an analgesic that can be harmful if used in excess. The synthesis of the film was conducted using a three-electrode system consisting of a gold (Au) electrode (working electrode), Ag/AgCl (reference electrode) and platinum electrode (counter electrode). In the first step, the gold electrode surface was modified with AuNPs. After that, the MIP film was deposited onto the modified electrode surface by electropolymerization. The MIP synthesis was performed by cyclic voltammetry, at a potential ranging from 0 to 1.3 V, at a scan rate of 100 mV/s for 30 scan cycles. The prepared electrodes were analyzed by CV and EIS. The performance of the MIP sensor was also studied by square wave voltammetry (SWV), in a concentration range between 4.5 ×10−5 and 5 × 10−7 M, resulting in a limit of detection of 2.3 × 10−9 M [72].
Another concerning problem is the side effect related to the use of dimetridazole beyond the permitted limits. Recently, Ali et al. [73] depicted the preparation of a poly-arginine MIP based sensor that can be used for electrochemical detection of dimetridazole. In this study, the MIP film synthesis was carried out by CV, using a three-electrode system (glassy carbon electrode GCE–working electrode, Ag/AgCl electrode–reference electrode and platinum wire electrode–counter electrode) and a solution consisting of L-arginine, dimetridazole dissolved in PBS. The electropolymerization conditions consisted of a potential range between −2 and 2.2 V, a scan rate of 100 mV/s and 12 scan cycles. The recognition experiments were performed by differential pulse voltammetry (DPV), using solutions with different concentration (10−10 to 10−5 M). The limit of detection for such sensors was 0.1 nM, which represents a promising future for dimetridazole detection [73].
In the past few years, both the production and consumption of antibiotics increased, which has led to a rise in environmental pollution. In recent years, sensors for antibiotics have been studied in order to improve the quality of life. For example, Ayankojo and co-workers [74] developed a sensor based on MIP for erythromycin detection, using a screen-printed electrode with Au working electrode. The electropolymerization was performed under potentiostatic condition, meaning a constant potential of 0.63 V, using m-phenylenediamine as monomer. The recognition and the rebinding capacity of the sensors were studied by DPV, pointing to a limit of detection of 0.1 nM and also a good selectivity. The results presented in this entry may represent a solution for erythromycin detection in aqueous solutions [74]. Another research group prepared an electrochemical sensor for tetracycline detection [75]. Theywere able tosynthesize an MIP film by electropolymerization on the surface of the gold electrode surface. A three-electrode system consisting of Au electrode (working electrode), saturated calomel electrode (reference electrode) and a platinum (Pt) electrode (counter electrode), was used in the electropolymerization process. A first step involved in synthesizing gold nanoparticles, further used in the development of the sensor. The films were prepared by CV upon applying a potential between 0.35 and 0.8 V, at a scan rate of 100 mV/s, for 10 scan cycles, after which the conditions were changed to a fixed potential (0.8 V). The performance of the sensor was studied by linear sweep voltammetry (LSV) and indicated quite high sensitivity, down to 0.22 fM tetracyclinein aqueous solutions [75].
Given the toxic effect of dyes on human health, controlling their presence in the food becomes mandatory. Thus, Arvand et al. [76] proposed the development of an electrochemical sensor based on an MIP layer that can be used for food analysis. In this case, the researchers used a three-electrode system consisting of a working electrode (functionalized GCE), reference electrode (saturated Ag/AgCl electrode) and counter electrode (Pt electrode). MWCNTs were used for the functionalization of the GCE. The electropolymerization was carried out by CV, in a potential range between −1.4 V and −0.4 V, for 15 scan cycles, using a solution containing AAm as the functional monomer, N, N-methylene-bis-acrylamide (MBA) as the crosslinker, sodium persulfate as the initiator, sunset yellow as the template molecule and sodium nitrate as the electrolyte dissolved in PBS. Ultimately, the sensor was electrochemically tested using CV and was demonstrated to possess good recognition properties for sunset yellow in the 0.05–100 µM concentration range and a low limit of detection (LOD) of 5 nM [76].
Shen et al. [77] described the preparation of an electrochemical sensor based on MIP for tetra-bromo-bisphenol A (TBBPA) detection. TBBPA is widely used for plastics and electronics manufacturing, and it may have negative effects on human health, including neurological and thyroid dysfunctions. Thus, the MIP films were prepared by electropolymerization onto GCE surface, using CV, in a potential range between −0.5 and 0.5 V, at 70 mV/s for 10 scan cycles. The solution used for electropolymerization consisted of ethanol, dopamine, TBBPA (template molecule) and PBS. MIPs showed a good sensitivity for TBBPA, in the 1–50 nM concentration range. Moreover, the MIP films presented selectivity and an LOD of 0.27 nM. The results are promising and comparable with further HPLC applications [77].
Wang and co-workers [78] prepared an MIP based sensor on indium tin oxide (ITO) bare for the determination of resveratrol. In this respect, prior to electropolymerization, the ITO electrode was modified with Ag nanoparticles using cyclic voltammetry and after that with a HAuCl4 (chloroauric acid) solution. Following this procedure, the MIP film was deposited onto the modified electrode surface via electropolymerization, by CV, applying a potential between 0 and 0.8 V at a scan rate of 50 mV/s, for 30 scan cycles. The electrochemical behavior of the sensors was studied by CV and EIS, in which case a linear response between 2.0 ×10−11 to 9.0 ×10−9 M was reported. The detection limit was determined to be 7.1 × 10−12 M. Moreover, the sensor was tested using structural analogues and proved good selectivity [78].
Li [79] described the development of an MIP sensor for atrazine detection. The MIP film synthesis was carried out by electropolymerization using a classical electrochemical cell. For producing the MIP film, the electrochemical cell consisted of an Au electrode (working electrode), a saturated calomel electrode (reference electrode), a Pt electrode (counter electrode), o-Phenylenediamine (monomer), electrolyte and atrazine (template molecule). In this case, the employed electropolymerization conditions referred to applying a potential between 0 and 0.8 V at a scan rate of 50 mV/s for 15 scan cycles. CV and DPV assays have proven that the prepared sensor can detect atrazine at different concentration, having a very low LOD of 1 × 10−9 M [79].
Motia et al. proposed an electrochemical sensor based on an MIP layer for sodium lauryl sulfate (SLS) detection. In this study, a “3 in 1” electrode with Au electrode as working electrode, Ag electrode as reference electrode and gold strip plate as counter electrode, was used. The MIP film was deposited by electropolymerization of 2-Aminothiophenol (2-ATP), in the presence of SLS (template molecule), by applying a potential between −0.35 and 0.80 V for 10 scan cycles at 100 mV/s. Prior to electropolymerization, the surface of the electrode was modified with a layer of 2-ATP by dripping the solution on the electrode surface. The electrochemical response, by CV, DPV and EIS, indicated that the sensor presented good affinity for the SLS, with a limit of detection of 0.18 pg/mL [80].
Seguro et al. [81] have recently present an electrochemical MIP-based sensor for diclofenac detection. The development of the voltametric sensor involved the synthesis of an MIP film by CV directly on screen printed carbon electrode (SPCE) surface, using a solution containing dopamine as monomer, diclofenac sodium as template molecule and KCl as electrolyte. The electrodeposition was performed in a potential range between −0.5 V and 1 V at a scan rate of 100 mV/s. Ultimately, the sensor was tested by DPV, and, according to the researchers, the imprinting factor was 2.5. Furthermore, a LOD of 70 nM and a limit of quantification (LOQ) of 200 nM were obtained [81].
Another sensor based on Au nanoparticles and MIPs was introduced by Liu and her group [82] for epinephrine detection. The researchers used a three-electrode system consisting of GCE (working electrode), saturated calomel electrode (reference electrode) and a Pt electrode (counter electrode). The working electrode was modified previously with HAuCl4, after which the electrodeposition was performed by applying a potential between −0.2 and 1.2 V for 20 scan cycles in the presence of epinephrine and 3-thiophene boronic acid (3-TBA). CV and DPV techniques were used to evaluate the performance of the final sensor, in which case a detection limit of 7.6 × 10−8 M was recorded [82].
Lopes and his group [83] proposed a sensor for naloxone detection. In this respect, the MIP film was synthesized by electropolymerization on the screen-printed carbon electrode that was firstly modified with MWCNT. The MIP film was prepared by CV when applying a potential in a range between −0.2 and 1 V, for 20 scan cycles at a scan rate of 100 mV/s, using a solution of 4-aminobenzoic acid and naloxone in PBS. The performance of the sensor was further studied using DPV and a detection limit of 0.2 µM was acquired [83]. Three years later, Shaabani and co-workers [84] also studied a sensor for naloxone detection, whereas the MIP film was deposited under similar conditions as described by Lopes et al. [83]. Yet, in this case, the SPCE surface was modified with Au nanoparticles and the CVs were obtained after applying a potential between −0.2 and 1 V at a scan rate of 50 mV/s for 15 scan cycles. By doing so, a lower detection limit (0.16 µM) was achieved.
Methylone is a synthetic drug, which produces psychotropic effects similar to ecstasy. Considering the side effects of this drug that include hypertension, paranoia and tachycardia, Couto et al. [85] presented a method for preparing an MIP-based sensor for methylone detection. The film was deposited on screen-printed gold electrode by electropolymerization of 2-mercaptobenzimidazole (monomer) in presence of the template molecule (methylone). The process was performed by CV when applying a potential in a range between −0.2 and 1.3 V for 15 scan cycles. A good performance of the sensor was obtained by SWV, in which case a detection limit of 1.1 µM was obtained. Moreover, the sensor also showed good stability and selectivity [85].
Melamine is a synthetic compound that can be used in the plastic industry as well as the milk industry. Considering the side effects of the melamine on human health (e.g., kidney problems, kidney stones), the monitoring of food is required. In this respect, Li and colleagues [86] developed an SPR sensor based on MIP films for melamine detection. The three-electrode system consisted of an optical fiber probe coated with Cr/Au film (working electrode), an Ag/AgCl electrode (reference electrode) and Pt electrode (counter electrode), while the MIP film was synthesized by electropolymerization of o-aminophenol (functional monomer) in the presence of melamine, by applying a potential between −0.3 and 1.2 V for 30 scan cycles at 50 mV/s. In order to estimate the performance of the sensor, several melamine solutions with different concentrations were used, leading to an LOD of 5.1 × 10−12 M, which indicated a good sensitivity of the sensor [89]. Two years later, Regasa et al. [87] proposed a sensor for melamine detection that involved the electropolymerization of aniline in order to obtain the MIP film. In this case, a three-electrode system with GCE (working electrode), Pt electrode (counter electrode) and Ag/AgCl electrode (reference electrode) was employed and the electrodeposition of the MIP film was carried out in potentiodynamic conditions, by applying a potential between −0.2 and 1 V for 10 scan cycles. This group [87] evaluated the properties of prepared sensors by CV and SWV, in which case they obtained a higher detection limit (4.47 × 10−10 M) compared to the previous work [86].
Roushani and his team [88] developed an MIP sensor for electrochemical detection of asulam herbicide or methyl N-(4-aminophenylsulfonyl)carbamate. In the first step, the GCE was modified with g-C3N4 (synthesized powder) and, subsequently, used for the electrodeposition of the MIP. The MIP solution consisted of monomer (dopamine), the template molecule (asulam) and electrolyte (KCl) dissolved intris-buffered saline (TBS) (pH = 7), which was electropolymerized in the −0.5 to 0.5 V potential range, for 10 cycles. The electrochemical behavior was analyzed using different methods, including CV, DPV and EIS, which led to the conclusion that the sensors possess high selectivity and affinity towards asulam, with an LOD close to 0.17 pM [88].
Wei and colleagues [89] studied and developed a sensor based on MIP for electrochemical detection of luteolin, using a three-electrode system (ITO as the working electrode, SCE as a reference electrode and Pt electrode as counter electrode). The MIP film synthesis was achieved by electropolymerization of β-cyclodextrin (monomer) in the presence of luteolin, applying a potential between −0.8 and 1.1 V for 20 cycles. Rebinding and selectivity experiments were performed by contacting the sensor with luteolin, respectively with quercetin and apigenin solutions and studied by DPV. In this case, the sensor detection limit for luteolin was near 2.4 × 10−8 M [89].
Zheng and co-workers [90] proposed a two-step fabrication process of an electrochemical MIP-based sensor for detection of 4-nonylphenol (4-NP) in milk samples. In the first step, a multilayer electrode was obtained using reduced graphene and HAuCl4. In the second step, the MIP film was prepared by electropolymerization of p-aminothiophenol (monomer), in presence of 4-NP as template molecule, upon applying a potential between −1 and 1 V. CV and DPV were employed for determining the sensor characteristics, in which case indicated a good selectivity and a LOD of 0.01 ng/mL [90].
The study by Mathieu-Scheers and her group [91] presented an MIP sensor for electrochemical detection of anthracene using GCE. The procedure was performed in a potential range between 0 and 1.4 V or 0 and 1.2 V, at a scanning rate of 10 mV/s, for 5 scan cycles, using pyrrole as a monomer and anthracene as a template molecule. The rebinding of anthracene was detected by SWV when a detection limit of 12 nM was calculated [91].
Recently, Grothe et al. [92] proposed a sensor based on MIP that allowed for detecting and identifying components from cocaine samples. Identification of the components provides important information regarding the origin of the cocaine. Herein, the MIP film was synthesized by electropolymerization of 3-amino-4-hydroxybenzoic in the presence of anesthetic benzocaine as the template molecule, on the carbon electrode of a SPCE. The electropolymerization process was performed in a potential range between 0 and 1.5 V for 10 scan cycles at a scan rate of 100 mV/s. SWV and EIS were used to study the electrochemical behavior of the films after contact with artificial urine (containing benzocaine) or caffeine, aminopyrine and procaine, in which case a limit of detection of 2.9 nM was achieved [92].
Radi and co-workers [93] have also described a sensor developed using MIP films that can be used for entacapone detection. A three-electrode system involving a WE (GCE), CE (platinum wire electrode) and RE (Ag/AgCl/KCl) was employed herein, while the MIP layers were prepared using polyphenylenediamine (monomer) and entacapone (template). Electropolymerization was performed using CV by applying a potential between 0 and 0.8 V at a scan rate of 100 mV/s and electrochemical behavior of the films was studied using EIS and DPV. As a result of the study, the sensor presented a good selectivity for entacapone vs. levodopa and carbidopa, with a limit of detection near 5 × 10−8 M [93].
