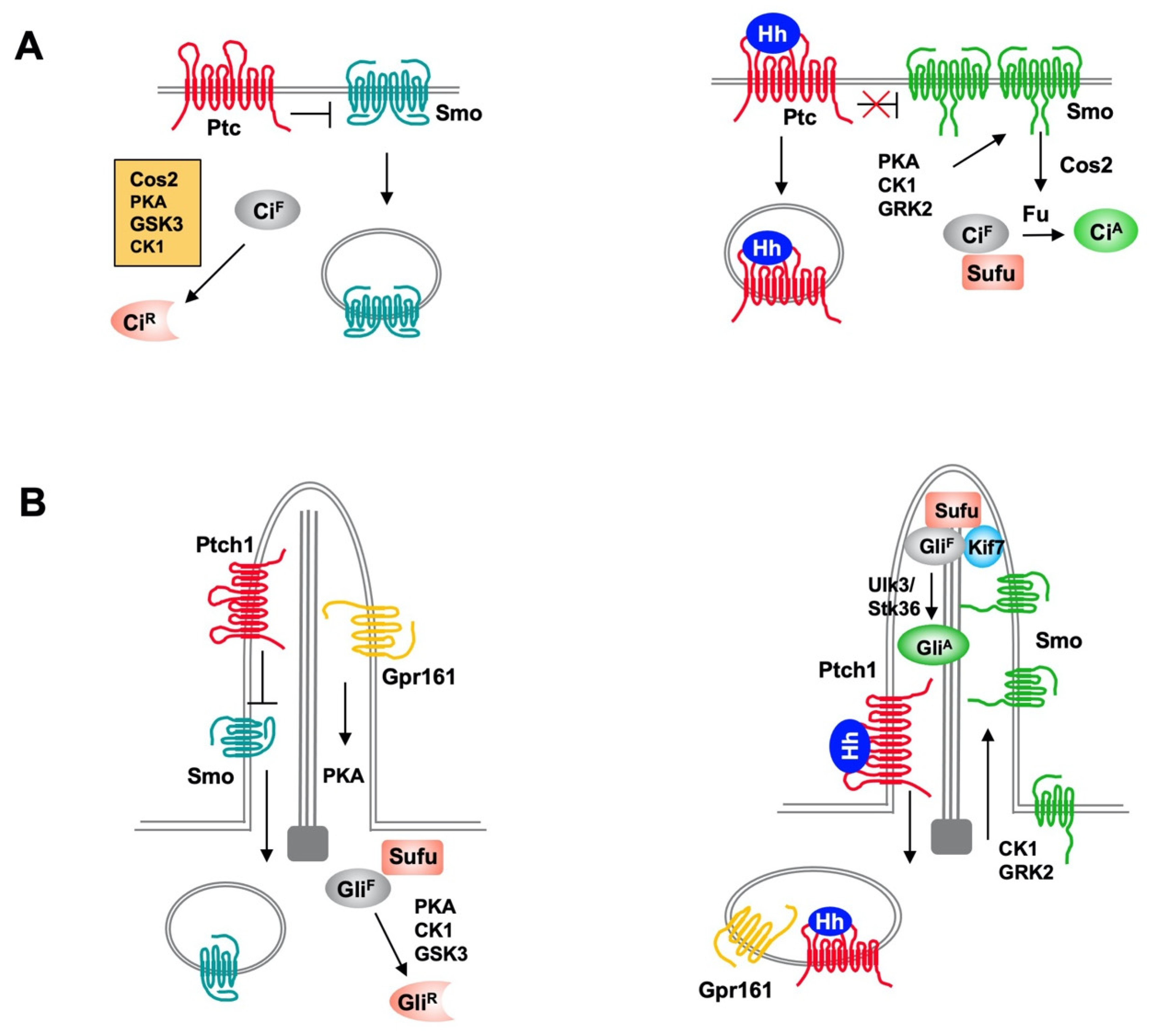
The Hedgehog (Hh) family of secreted proteins governs embryonic development and adult tissue homeostasis in species ranging from insects to mammals. Deregulation of Hh pathway activity has been implicated in a wide range of human disorders, including congenital diseases and cancer. Hh exerts its biological influence through a conserved signaling pathway. Binding of Hh to its receptor Patched (Ptc), a twelve-span transmembrane protein, leads to activation of an atypical GPCR family protein and Hh signal transducer Smoothened (Smo), which then signals downstream to activate the latent Cubitus interruptus (Ci)/Gli family of transcription factors. Hh signal transduction is regulated by ubiquitination and deubiquitination at multiple steps along the pathway including regulation of Ptc, Smo and Ci/Gli proteins.
- hedgehog
- ubiquitination
- deubiquitination
1. General Introduction
2. Regulation of Hedgehog Signal Transduction by Ubiquitination and Deubiquitination
2.1. Regulation of Hh Pathway Transcription Factors Ci/Gli by Ubiquitination and Deubiquitination
Hh signaling controls the balance between the repressor and activator forms of the Ci/Gli family of transcription factor. In the absence of Hh, full-length Ci/Gli is proteolytically processed to form truncated repressor (CiR/GliR) that lacks the C-terminal coactivator domain but retains the N-terminal corepressor domain. Hh signaling blocks the proteolysis of Ci/Gli and converts accumulated full-length Ci/Gli into activator form CiA/GliA. Both the production of CiR/GliR and abundance of CiA/GliA are regulated by the ubiquitin and proteasome system (UPS) (Figure 2) [8][22].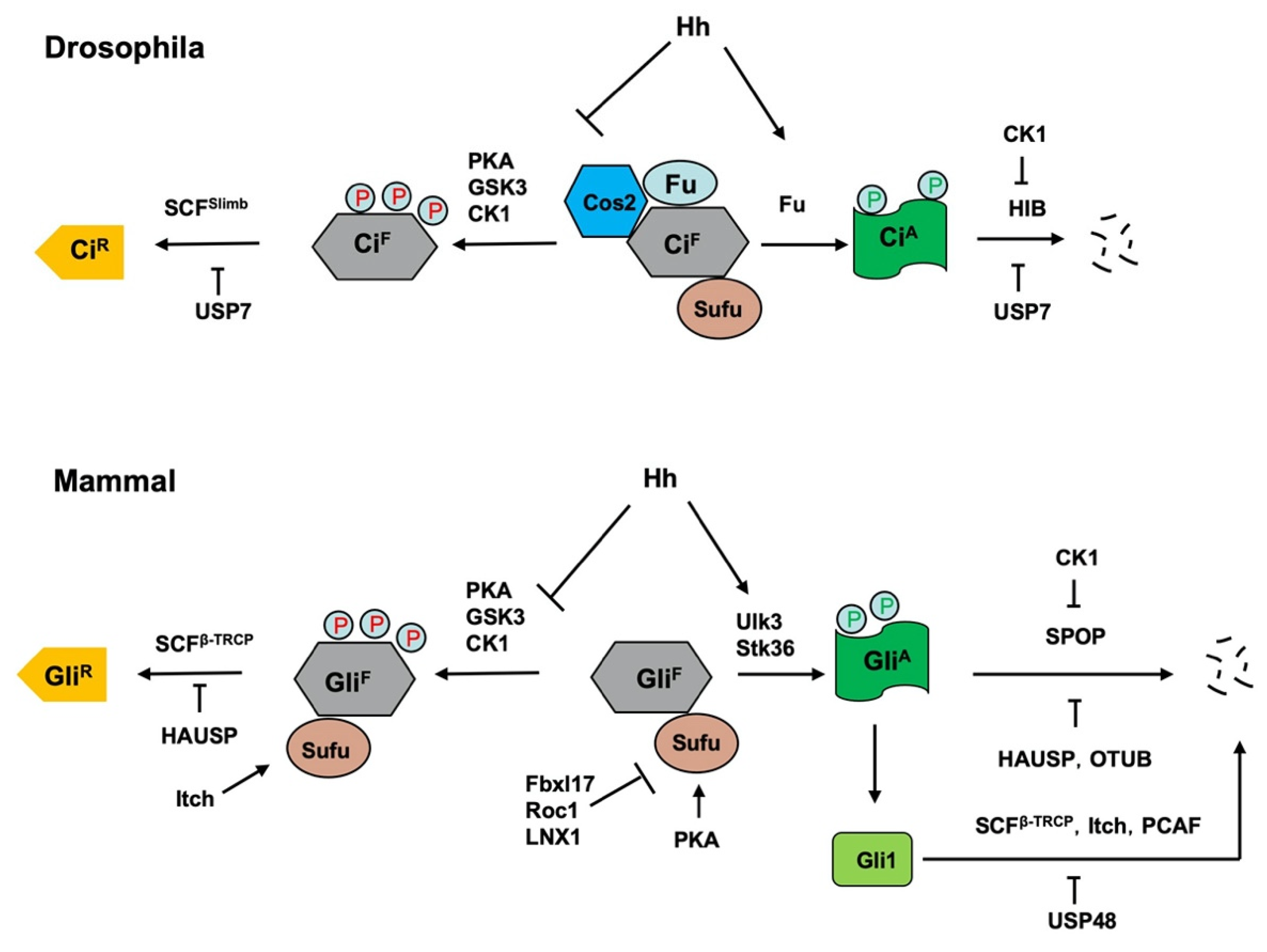 Figure 2. Regulation of Ci/Gli processing and degradation by E3s and DUBs. Hh signaling controls the balance between the repressor and activator forms of the Ci/Gli, which is regulated by the ubiquitin and proteasome system (UPS). In the absence of Hh, full-length Ci/Gli (mainly Gli3) is proteolytically processed to form truncated repressor (CiR/GliR) through PKA/GSK3/CK1-mediated phosphorylation, followed by SCFSlimb/SCFβ-TRCP-mediated ubiquitination. The DUB USP7/HAUSP antagonizes this process through deubiquitinating Ci/Gli. The E3 ligase Itch promotes GliR production by ubiquitinating Sufu to increase its interaction with Gli3. Hh signaling blocks the proteolysis of Ci/Gli and converts accumulated full-length Ci/Gli into activator form CiA/GliA though phosphorylation by the Fu family kinases. When dissociated from Sufu, CiA/GliA becomes labile and is degraded by HIB/SPOP, a process that is attenuated by CK1-meidated phosphorylation of Ci/Gli. The DUBs USP7/HAUSP and OTUB deubiquitinate Ci/Gli to inhibit its degradation. The E3 ligases SCFβ-TRCP, Itch and PCAF promote Gli1 degradation while the DUB USP48 antagonizes this process. In addition, mammalian Sufu is degraded by multiple E3 ligases including Fbxl17, ROC1 and LNX1.
Figure 2. Regulation of Ci/Gli processing and degradation by E3s and DUBs. Hh signaling controls the balance between the repressor and activator forms of the Ci/Gli, which is regulated by the ubiquitin and proteasome system (UPS). In the absence of Hh, full-length Ci/Gli (mainly Gli3) is proteolytically processed to form truncated repressor (CiR/GliR) through PKA/GSK3/CK1-mediated phosphorylation, followed by SCFSlimb/SCFβ-TRCP-mediated ubiquitination. The DUB USP7/HAUSP antagonizes this process through deubiquitinating Ci/Gli. The E3 ligase Itch promotes GliR production by ubiquitinating Sufu to increase its interaction with Gli3. Hh signaling blocks the proteolysis of Ci/Gli and converts accumulated full-length Ci/Gli into activator form CiA/GliA though phosphorylation by the Fu family kinases. When dissociated from Sufu, CiA/GliA becomes labile and is degraded by HIB/SPOP, a process that is attenuated by CK1-meidated phosphorylation of Ci/Gli. The DUBs USP7/HAUSP and OTUB deubiquitinate Ci/Gli to inhibit its degradation. The E3 ligases SCFβ-TRCP, Itch and PCAF promote Gli1 degradation while the DUB USP48 antagonizes this process. In addition, mammalian Sufu is degraded by multiple E3 ligases including Fbxl17, ROC1 and LNX1.
2.2. Regulation of Hh Intracellular Signaling Components by Ubiquitination
The intracellular Hh signal transduction pathway contains several conserved signaling components including the kinesin-like protein Cos2/Kif7, the Fu family kinases Fu/Ulk3/Stk36, and the tumor suppressor Sufu [3]. In Drosophila, Cos2 and Fu form a complex to promote Ci phosphorylation, ubiquitination and proteolytic processing to form CiR in the absence of Hh [13]. In response to Hh stimulation, Cos2 recruits Fu to activated Smo to induce Fu dimerization/oligomerization, autophosphorylation and activation [23][24]; activated Fu then phosphorylates Ci to convert it into CiA by altering its binding to Sufu [17]. In mammalian cells, Kif7 also plays a dual role in regulating the formation of both GliR and GliA [25][26][27]; however, the underlying mechanisms remain obscured although Kif7 has been shown to organize the cilium tip compartment where Gli is thought to be activated [28][29]. The Fu family kinases Ulk3 and Stk36 play a partially redundant role in phosphorylating and activating Gli proteins by alleviating Sufu-mediated repression [17]. Several studies revealed that both Cos2/Kif7 and Sufu are regulated by the UPS.3. Conclusions
References
- Ingham, P.W.; McMahon, A.P. Hedgehog signaling in animal development: Paradigms and principles. Genes Dev. 2001, 15, 3059–3087.
- Jiang, J.; Hui, C.C. Hedgehog signaling in development and cancer. Dev. Cell 2008, 15, 801–812.
- Jiang, J. Hedgehog signaling mechanism and role in cancer. Semin. Cancer Biol. 2021.
- Iriana, S.; Asha, K.; Repak, M.; Sharma-Walia, N. Hedgehog Signaling: Implications in Cancers and Viral Infections. Int. J. Mol. Sci. 2021, 22, 1042.
- Chen, Y.; Jiang, J. Decoding the phosphorylation code in Hedgehog signal transduction. Cell Res. 2013, 23, 186–200.
- Ma, G.; Li, S.; Han, Y.; Li, S.; Yue, T.; Wang, B.; Jiang, J. Regulation of Smoothened Trafficking and Hedgehog Signaling by the SUMO Pathway. Dev. Cell 2016, 39, 438–451.
- Li, S.; Li, S.; Han, Y.; Tong, C.; Wang, B.; Chen, Y.; Jiang, J. Regulation of Smoothened Phosphorylation and High-Level Hedgehog Signaling Activity by a Plasma Membrane Associated Kinase. PLoS Biol. 2016, 14, e1002481.
- Jiang, J. Regulation of Hh/Gli signaling by dual ubiquitin pathways. Cell Cycle 2006, 5, 2457–2463.
- Liu, A. Proteostasis in the Hedgehog signaling pathway. Semin. Cell Dev. Biol. 2019, 93, 153–163.
- Bufalieri, F.; Lospinoso Severini, L.; Caimano, M.; Infante, P.; Di Marcotullio, L. DUBs Activating the Hedgehog Signaling Pathway: A Promising Therapeutic Target in Cancer. Cancers 2020, 12, 1518.
- Zhang, J.; Liu, Z.; Jia, J. Mechanisms of Smoothened Regulation in Hedgehog Signaling. Cells 2021, 10, 2138.
- Wilson, C.W.; Chuang, P.T. Mechanism and evolution of cytosolic Hedgehog signal transduction. Development 2010, 137, 2079–2094.
- Zhang, W.; Zhao, Y.; Tong, C.; Wang, G.; Wang, B.; Jia, J.; Jiang, J. Hedgehog-regulated costal2-kinase complexes control phosphorylation and proteolytic processing of cubitus interruptus. Dev. Cell 2005, 8, 267–278.
- Jia, J.; Zhang, L.; Zhang, Q.; Tong, C.; Wang, B.; Hou, F.; Amanai, K.; Jiang, J. Phosphorylation by double-time/CKIepsilon and CKIalpha targets cubitus interruptus for Slimb/beta-TRCP-mediated proteolytic processing. Dev. Cell 2005, 9, 819–830.
- Smelkinson, M.G.; Kalderon, D. Processing of the Drosophila hedgehog signaling effector Ci-155 to the repressor Ci-75 is mediated by direct binding to the SCF component Slimb. Curr. Biol. 2006, 16, 110–116.
- Qi, X.; Schmiege, P.; Coutavas, E.; Wang, J.; Li, X. Structures of human Patched and its complex with native palmitoylated sonic hedgehog. Nature 2018, 560, 128–132.
- Han, Y.; Wang, B.; Cho, Y.S.; Zhu, J.; Wu, J.; Chen, Y.; Jiang, J. Phosphorylation of Ci/Gli by Fused Family Kinases Promotes Hedgehog Signaling. Dev. Cell 2019, 50, 610–626.
- Bangs, F.; Anderson, K.V. Primary Cilia and Mammalian Hedgehog Signaling. Cold Spring Harb. Perspect. Biol. 2017, 9, a028175.
- Kong, J.H.; Siebold, C.; Rohatgi, R. Biochemical mechanisms of vertebrate hedgehog signaling. Development 2019, 146, dev166892.
- Corbit, K.C.; Aanstad, P.; Singla, V.; Norman, A.R.; Stainier, D.Y.; Reiter, J.F. Vertebrate Smoothened functions at the primary cilium. Nature 2005, 437, 1018–1021.
- Rohatgi, R.; Milenkovic, L.; Scott, M.P. Patched1 regulates hedgehog signaling at the primary cilium. Science 2007, 317, 372–376.
- Jiang, J. Degrading Ci: Who is Cul-pable? Genes Dev. 2002, 16, 2315–2321.
- Shi, Q.; Li, S.; Jia, J.; Jiang, J. The Hedgehog-induced Smoothened conformational switch assembles a signaling complex that activates Fused by promoting its dimerization and phosphorylation. Development 2011, 138, 4219–4231.
- Zhang, Y.; Mao, F.; Lu, Y.; Wu, W.; Zhang, L.; Zhao, Y. Transduction of the Hedgehog signal through the dimerization of Fused and the nuclear translocation of Cubitus interruptus. Cell Res. 2011, 21, 1436–1451.
- Cheung, H.O.; Zhang, X.; Ribeiro, A.; Mo, R.; Makino, S.; Puviindran, V.; Law, K.K.; Briscoe, J.; Hui, C.C. The kinesin protein Kif7 is a critical regulator of Gli transcription factors in mammalian hedgehog signaling. Sci. Signal. 2009, 2, ra29.
- Endoh-Yamagami, S.; Evangelista, M.; Wilson, D.; Wen, X.; Theunissen, J.W.; Phamluong, K.; Davis, M.; Scales, S.J.; Solloway, M.J.; de Sauvage, F.J.; et al. The mammalian Cos2 homolog Kif7 plays an essential role in modulating Hh signal transduction during development. Curr. Biol. 2009, 19, 1320–1326.
- Liem, K.F., Jr.; He, M.; Ocbina, P.J.; Anderson, K.V. Mouse Kif7/Costal2 is a cilia-associated protein that regulates Sonic hedgehog signaling. Proc. Natl. Acad. Sci. USA 2009, 106, 13377–13382.
- He, M.; Subramanian, R.; Bangs, F.; Omelchenko, T.; Liem, K.F., Jr.; Kapoor, T.M.; Anderson, K.V. The kinesin-4 protein Kif7 regulates mammalian Hedgehog signalling by organizing the cilium tip compartment. Nat. Cell Biol. 2014, 16, 663–672.
- Han, Y.; Xiong, Y.; Shi, X.; Wu, J.; Zhao, Y.; Jiang, J. Regulation of Gli ciliary localization and Hedgehog signaling by the PY-NLS/karyopherin-beta2 nuclear import system. PLoS Biol. 2017, 15, e2002063.
- Kim, J.; Hsia, E.Y.; Brigui, A.; Plessis, A.; Beachy, P.A.; Zheng, X. The role of ciliary trafficking in Hedgehog receptor signaling. Sci. Signal. 2015, 8, ra55.
