Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Rita Xu and Version 1 by Dabing Zhang.
The actin cytoskeleton is crucial for plant morphogenesis, and organization of actin filaments (AF) is dynamically regulated by actin-binding proteins. However, the roles of actin-binding proteins, particularly type II formins, in this process remain poorly understood in plants.Here, we report that a type II formin in rice, Oryza sativa formin homolog 3 (OsFH3), acts as a major player to modulate AF dynamics and contributes to rice morphogenesis.
- OsFH3
- morphological defects
- profilin–actin complex
1. Introduction
Microfilaments and microtubules are two important cytoskeletal components in plant cells. Microfilaments are actin filaments (AF) composed of polymerized globular actin (G-actin) monomers. The initial nucleation of monomers to form a new AF, and subsequent AF elongation, bundling of filaments into cables, and cross-interaction of cables are all mediated by actin-binding proteins. In vivo, actin monomers bind to the small protein profilin, which inhibits spontaneous nucleation and elongation to sustain homeostasis, and serves as a nucleotide exchange factor for actin, increasing the exchange of ATP or ADP at least fivefold [1,2][1][2].
The AF network in plants plays indispensable roles in many biological processes, including morphogenesis [3], signal transduction [4,5][4][5], stomatal opening and closing [6], hormone signaling [7[7][8],8], immunity [9[9][10],10], organelle transport [11[11][12],12], cell division [13], and cell growth [14]. Several classes of actin-binding proteins have been implicated in AF formation, including formins, actin-related proteins-2/3 (Arp2/3), capping proteins, and enabled/vasodilator-stimulated phosphoproteins (Ena/VASP) [15,16,17,18,19][15][16][17][18][19].
Formins in higher plants are divided into two types according to their N-terminal domains [20,21][20][21]. Type I formins contain a transmembrane domain and are associated with cell wall proteins [21]; type II formins contain a phosphatase and tensin homolog (PTEN) domain essential for interaction with membranes [3,20,22][3][20][22].
Both types contain C-terminal FH1 and FH2 domains, which are evolutionarily conserved [23,24][23][24]: the FH1 domain promotes AF elongation by binding to profilin–actin monomers via its polyproline domains [25[25][26],26], while the FH2 domain is essential for AF nucleation [3,27][3][27].
Compared with the known functions of type I formins, for example, in affecting pollen tube growth [27], cytokinesis [28], membrane trafficking pathways [29], and root cell ontogeny [30], the function of type II formins is largely unknown. Single knockouts of formin genes in plants generally exhibit only mild phenotypes, likely due to functional redundancy between formin family proteins [31,32,33,34][31][32][33][34]. Knockouts of two type II formins have been reported in moss; while single mutants do not show any obvious phenotype, double mutants are severely stunted with disrupted actin organization [35].
The model dicot plant Arabidopsis thaliana has 21 predicted formins, including 10 type II members. Among characterized type II formins, AtFH14 is involved in AF-microtubule interactions, regulating their dynamics during cell division [36], while AtFH13 and its distant homolog AtFH14 form heterodimers to associate with microtubules and the periphery of the endoplasmic reticulum [37].
The model monocot rice has five type II formins. Among them, only OsFH5 has been functionally characterized; OsFH5 acts as a core regulator of actin nucleation and AF elongation and associates with the microtubule network [3,38][3][38]. Loss-of-function osfh5 mutants exhibit serious defects in vegetative and reproductive growth [3], such as delayed cell growth [39], stronger sensitivity to gravity in root [40], and abnormal shoot gravitropism [41]. In addition, OsFH15, a rice type I formin, interacts with both AF and microtubules to regulate grain size by affecting cell expansion [33].
2. Phenotypes of osfh3 Mutant
To understand the function of OsFH3, we used the 2 kb OsFH3 promoter to drive the expression of the β-glucuronidase (GUS) reporter gene. GUS signals were detected in root, stem, leaf, and pedicle but not in root tips and anthers. The ubiquitous expression profile of OsFH3 is, thus, consistent with those of most reported rice formins [38]; however, it is discrete from that of reported type II formin gene OsFH5, whose expression is observed in root tips but not in pedicle [3].
To determine the molecular function of OsFH3, two loss-of-function osfh3 mutants were generated using CRISPR/Cas9 targeting in the second exon of the N-terminal PTEN domain, which caused premature protein truncation in wild-type (WT) plants. Because of OsFH3 and OsFH5, the two of the known five type II formins in rice [42], four CRISPR/Cas9-induced osfh3 mutants (targeting the same sequence) were generated in an available osfh5 line (also called rice morphology determinant1-1, rmd1-1 [3]). These resulting osfh3osfh5 double mutants were then used to investigate their functional redundancy. The osfh3 mutant was semi-dwarf, while the osfh5 and osfh3 osfh5 mutants exhibited more severe phenotypes: dwarf with smaller seeds and curved roots (Figure 1A).


Figure 1. Phenotypes of WT, osfh3, and osfh5 single and double mutant plants. (A) Plant height at grain maturation. Bar = 10 cm. (B) Mature grains. Bar = 1 cm. (C–F) Lateral roots of 3-day old seedlings. Bar = 1 mm.
Other phenotypes, such as seed length, panicle primary branch numbers, and root hair curling, were affected by OsFH3 and OsFH5 independently. Compared with WT plant, mutants exhibited shorter seed length and reduced primary branches in the inflorescence: seed length of the single and double mutant was about 94% and 73% of that of WT plant, respectively (Figure 1B); and the number of primary branches in the inflorescence of the single and double mutant was about 80% and 58% of that of WT plant, respectively. Compared with WT and osfh3 or osfh5 single mutants, lateral roots of osfh3 osfh5 double mutants were much curlier (Figure 1C–F). These results indicate that both OsFH3 and OsFH5 play important roles in regulating rice morphogenesis and that knockout of OsFH3 exacerbates the phenotypic defects in the osfh5 mutant background.
To test the cellular state of roots in mutants, propidium iodide (PI) was used (Figure 2A–E). The staining results showed that changes in cell numbers between the inside and the outside walls of curved roots were not significant (Figure 2F), while the cell length located inside of the curved roots became shorter than that located outside of the curved roots in osfh3 osfh5 mutant lines (Figure 2D,G). However, such a cell length difference between the inside and the outside wall of roots was not observed in other plants, including WT, osfh3, and osfh5. These results indicated that root curvature observed in osfh3 osfh5 is caused by changes in the length but not the number of cells on the opposite side of the root.
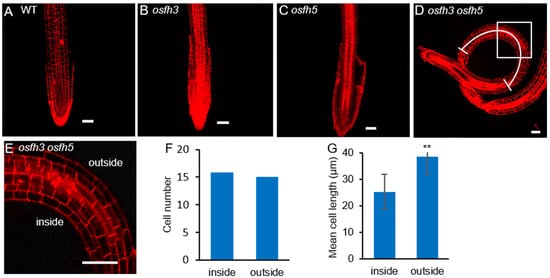

Figure 2. Propidium iodide staining results of WT, osfh3, osfh5, and osfh3 osfh5 roots tips. (A–D) WT, osfh3, osfh5, and osfh3 osfh5 roots tips were stained with propidium iodide. Bar = 50 µm. (E) Enlarged pictures of boxes in the D pictures. (F) Cell number statistic of curved segment in osfh3 osfh5 roots. n = 10. (G) Cell length statistic of curved segment in osfh3 osfh5 roots. The arc in D marks the data source area in the root. n = 100. ** p < 0.01, Student’s t-test.
Previous studies reported that osfh5 mutants are more sensitive to gravity [40], and this response is affected by sunlight [41]. Our results revealed that the sensitivity of osfh3 osfh5 roots to gravity is similar to that of osfh5 roots and that osfh3 roots do not show an obvious gravitropic phenotype. In addition, double mutant exhibited a different phenotype to light from either single mutant. osfh3 seedlings (5-d old) grew upright under both light and dark conditions, osfh5 seedlings (5-d old) grew curved under light but straight under dark conditions, while osfh3 osfh5 grew bent under both light and dark conditions. Thus, OsFH3 appears to function in gravity-sensing under dark, which is distinct from OsFH5.
3. OsFH3 Nucleates Actin
To obtain the mechanistic insights into the function of OsFH3 in rice morphology, Alexa Fluor 488®-phalloidin was used to stain AF in the lateral roots of 3-day-old seedlings (Figure 3A–D) to examine the effect of OsFH3 on actin nucleation and AF connection. While AF length was obviously reduced in osfh5 and even further reduced in osfh3 osfh5 lines, there was no obvious reduction in AF length of osfh3 compared with WT (Figure 3I). In addition, while decreased bundling value and increased AF abundance were observed in osfh3 (Figure 3J,K), such changes were not found in osfh5 or double mutant.
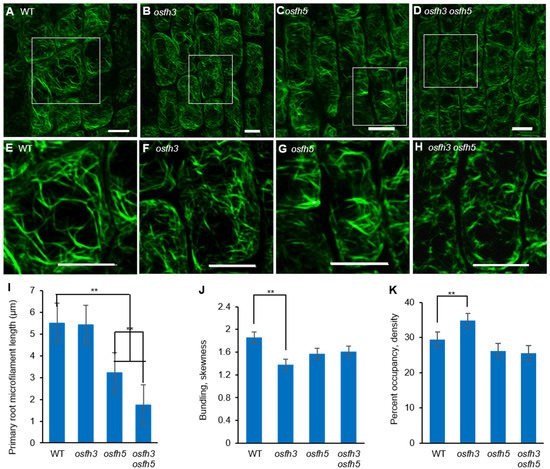

Figure 3. Microfilaments staining results with Alexa Fluor 488®-phalloidin. (A–D) Alexa Fluor 488®-phalloidin stained lateral root elongation zone of 3-day-old rice seedlings. Bar = 10 µm. (E–H) Enlarged pictures of white boxes in A to D, respectively. Bar = 10 µm. (I) Length of microfilaments in primary roots. Mean ± SD, n > 100 microfilaments from 10 cells across 5 plants. ** p < 0.001, one-way ANOVA. (J) Bundling (skewness) of 3-day-old seedling roots. Mean ± SD, n = 25 cells across 5 plants. ** p < 0.001, one-way ANOVA. (K) Percentage of occupancy (Actin filament abundance) of 3-day-old seedling roots. Mean ± SD, n = 25 cells across 5 plants. ** p < 0.001, one-way ANOVA. Region selection was divided by cells of pictures from elongation zone and quantified with ImageJ.
To further unravel the biological role of OsFH3, expressing the full length of OsFH3 in E. coli was performed to explore OsFH3′s function in actin assembly, which, unfortunately, failed to generate functional protein, likely due to the difficulty in expressing 15 highly repetitive polyproline segments within the FH1 domain. As an alternative approach, the truncated FH1–FH2 segment (containing four polyproline segments) and the FH2 domain were expressed and purified (Figure 4A). These protein segments were incubated in the presence of actin monomers to examine their effects on actin assembly. Both OsFH3 FH2 and OsFH3 FH1–FH2 decreased the initial lag of actin monomer association in a dose-dependent manner, indicating their active roles in actin nucleation (Figure 4A,B).
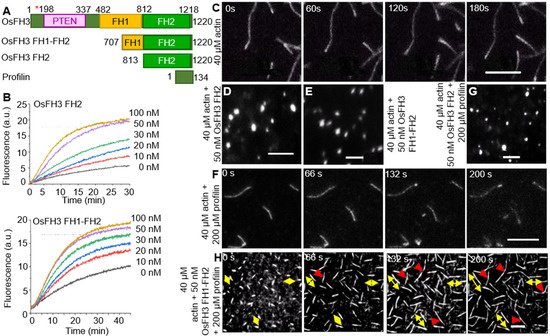

Figure 4. OsFH3 nucleates actin and promotes formation and connection of AF. (A) Schematic diagram of OsFH3 structure, indicating FH1–FH2 and FH2 constructs used for actin testing. Amino acid positions in the full-length protein are given. Red asterisk indicates mutation position in osfh3 mutants. PTEN, phosphatase, and tensin homolog domain. Profilin is PRF4 from Arabidopsis. (B) Time course of actin nucleation in the presence of OsFH3 FH2 (above) or OsFH3 FH1–FH2 (below). Pyrene fluorescence (a.u., arbitrary units) is proportional to nucleation. (C–E) The effect of OsFH3 domains on AF elongation visualized by time-lapse fluorescence microscopy. Fluorescence shows Oregon Green-labeled actin, showing AF elongation, in the actin-only experiment (C); and stalled elongation of AF (D,E) in the presence of OsFH3 FH2 and OsFH3 FH1–FH2 protein segments. Bar = 5 µm. (F–H) The effect of OsFH3 domains on AF elongation and bundling in the presence of profilin visualized by time-lapse fluorescence microscopy. Fluorescence shows Oregon Green-labeled actin. Actin and profilin only (F); actin, profilin, and OsFH3 FH2 (G); actin, profilin, and OsFH3 FH1–FH2 (H). Yellow arrows in H indicate bi-directional elongation of AF. Red arrows indicate connection of AF. Bar = 5 µm.
Time-lapse fluorescence microscopy was used to directly visualize AF elongation in the presence of OsFH3 protein segments. AF nucleation and elongation proceeded normally in the control experiment without the addition of OsFH3 proteins (Figure 4C). However, when OsFH3 FH2 or OsFH3 FH1–FH2 was added, small bright spots were observed, indicating that actin nucleation could occur, but elongation could not (Figure 4D,E). When profilin was added to the initial actin monomers, OsFH3 FH1–FH2 could promote AF elongation, but OsFH3 FH2 domain could not, likely due to its lack of polyproline sequences (Figure 4F–H). In the presence of profilin and OsFH3 FH1–FH2, the AF appeared to grow from both ends (Figure 4H, bi-directional yellow arrows), compared with only one direction of growth in the control experiment (Figure 4F). Moreover, when different AF extended to connecting points, the connecting ends of the AF ceased to elongate (Figure 4H, red arrows). These results indicate that OsFH3 protein is required for normal AF elongation and organization in the presence of the profilin–actin complex.
4. OsFH3 Bundles and Caps AF
To explore the effect of OsFH3 on AF binding and bundling, high- and low-speed co-sedimentation in vitro assays was performed using OsFH3 FH2 and OsFH3 FH1–FH2 proteins. In the high-speed assay, OsFH3 FH2 accumulated in the pellets in proportion to its initial concentration in solution, but only in the presence of actin (Figure 5A), indicating that OsFH3 FH2 can directly bind AF. In low-speed co-sedimentation, the amounts of AF in the pellet reached a maximum level with 2 μM OsFH3 FH2 protein (Figure 5B), demonstrating the bundling ability of OsFH3 FH2. The same results for both high- and low-speed assays were also observed for the OsFH3 FH1–FH2 protein, indicating that both OsFH3 FH2 and OsFH3 FH1–FH2 can bind and bundle AF.
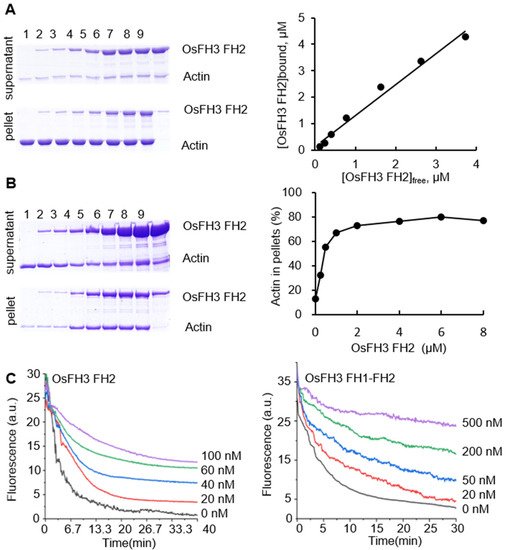

Figure 5. OsFH3 FH2 domain bundles and caps AF. (A) Determination of AF binding to OsFH3 FH2 using high-speed co-sedimentation assays. Lanes 1–8, 5 µM actin with 0, 0.25, 0.5, 1, 2, 4, 6, and 8 µM OsFH3 FH2, respectively; lane 9, 8 µM OsFH3 FH2, no actin. Graph (left) shows quantification of Coomassie staining on protein gel (right). (B) Determination of AF binding to OsFH3 FH2 using low-speed co-sedimentation assays. Lanes 1–8, 5 µM actin with 0, 0.25, 0.5, 1, 2, 4, 6, and 8 µM OsFH3 FH2, respectively; lane 9, 8 µM OsFH3 FH2, no actin. Graph (left) shows quantification of Coomassie staining on protein gel (right). (C) Kinetics of AF depolymerization in the presence of OsFH3 FH2 (left) or OsFH3 FH1–FH2 (right). Pyrene fluorescence (a.u., arbitrary units) is proportional to actin polymer concentration.
In view of other reports of formins’ capping ability [3[3][43],43], dilution-mediated AF depolymerization assays were performed. OsFH3 FH2 and OsFH3 FH1–FH2 segments were both observed to retard actin depolymerization in a dose-dependent manner (Figure 5C), suggesting that OsFH3 might cap and protect the barbed end of AF.
5. PTEN Domain Affects the Localization of OsFH3
An OsFH3-eGFP fusion protein was used to examine the subcellular localization of OsFH3 in transgenic rice lines and protoplasts. Punctate fluorescent signals were detected in the cytoplasm of rice coleoptile cells and protoplasts (Figure 6B,C). In order to figure out what these dot signals are and given the interaction between OsFH3 and AF, OsFH3-eGFP and the AF marker mScarlet-FABD2 were co-expressed in tobacco leaves, for the reason that co-expression of OsFH3-eGFP and AF marker experiment was failed and rice protoplasts could not clearly display the morphology of microfilaments and microtubules (rice protoplasts lost cell wall binding and vacuoles expand to squeeze the cytoplasm into a very small space). These results revealed that most OsFH3 colocalized with AF, predominantly at the intersections of the AF cytoskeleton (Figure 6D).
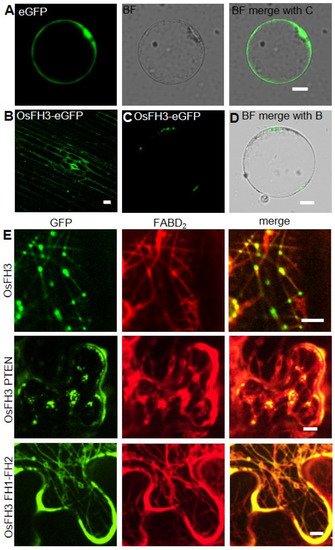

Figure 6. Colocalization of OsFH3 with AF in vivo. (A) Contrast, eGFP signals in rice protoplast. Bar= 10 µm. (B) OsFH3-eGFP signals in rice coleoptile cells. Bar = 10 µm. (C,D) OsFH3-eGFP signals in rice protoplast showing GFP (B) and merged bright field (BF) with GFP (C) images. Bar = 10 µm. (E) Colocalization of OsFH3-eGFP full length protein, OsFH3 PTEN-eGFP and OsFH3 FH1-FH2-eGFP domains with AF marker mScarlet-FABD2 in tobacco leaves. Bar = 5 µm.
Considering that the PTEN domains of type II plant formins guide protein localization and allow FH1-FH2 binding to AF [3], the OsFH3 protein was split into two halves, the N-terminal PTEN domain (aa 1 to 337) and the C-terminal FH1–FH2 domain (aa 551 to 1218), and each was fused to eGFP. When co-expressed with mScarlet-FABD2 in tobacco leaves, PTEN-eGFP co-localized with AF as the full-length protein did, but the AF became disordered (Figure 6E). The FH1–FH2-eGFP protein was also found to be localized across the whole length of highly ordered AF (Figure 6E). These results suggest that the PTEN domain plays a key role in OsFH3 accumulation at intersections of the AF network, while the FH1–FH2 domain helps to bind AF. Thus, PTEN and FH1–FH2 domains work together to determine the precise location of OsFH3 in vivo.
6. OsFH3 Binds Microtubules
Previous reports revealed that some formins, such as AtFH4, AtFH14 in Arabidopsis [37[37][44],44], and OsFH5 in rice [3], can bind and bundle microtubules. As OsFH3 is the homolog of OsFH5, the function for OsFH3 to bind and bundle microtubules was tested using high- and low-speed co-sedimentation in vitro assays with OsFH3 FH2 and OsFH3 FH1–FH2 domains. In the high-speed assays, slight increases of OsFH3 FH2 accumulation in pellets in the presence of microtubule were observed as the concentration of OsFH3 FH2 increased (Figure 7A,B), indicating that OsFH3 FH2 binding ability of microtubules is weaker than AF (Figure 5A,B). In low-speed assays, the content of microtubule and OsFH3 FH2 in the pellet was not positively related to the concentration of OsFH3 FH2 (Figure 7C,D), suggesting that OsFH3 cannot bundle microtubule. The same results for both high- and low-speed assays were also observed for the OsFH3 FH1–FH2 protein, indicating that both OsFH3 FH2 and OsFH3 FH1–FH2 can bind but not bundle microtubules.
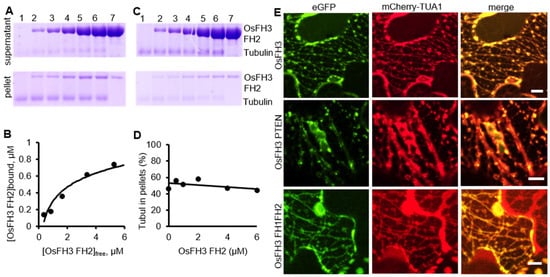

Figure 7. OsFH3 binds and colocalizes with microtubules. (A) High-speed co-sedimentation assays to determine tubulin binding to OsFH3 FH2. Lanes 1–6, 2 µM microtubule with 0.5, 1, 2, 4, and 6 µM OsFH3 FH2, respectively; lane 7, 6 µM OsFH3 FH2, no tubulin. (B) Graph shows quantification of Coomassie staining on protein gel (A). (C) Low-speed co-sedimentation assays to determine tubulin bundling with OsFH3 FH2. Lanes 1–6, 2 µM tubulin with 0.5, 1, 2, 4, and 6 µM OsFH3 FH2, respectively; lane 7, 6 µM OsFH3 FH2, no tubulin. (D) Graph shows quantification of Coomassie staining on protein gel (C). (E) Colocalization of OsFH3-eGFP full-length protein, PTEN, and FH1–FH2 domains with microtubule marker mCherry-TUA1 in tobacco leaves. Bar = 5 µm.
Next, OsFH3-eGFP, OsFH3 PTEN-eGFP, or OsFH3 FH1–FH2-eGFP was co-transfected with the microtubule marker mCherry-TUA1 into tobacco leaves to examine the in vivo colocalization. Full-length OsFH3 could colocalize with microtubule in a punctate distribution pattern (Figure 6E and Figure 7E), similar but not identical to the colocalization of OsFH3 with AF. PTEN-eGFP also co-localized with microtubules (Figure 7E), while FH1FH2-eGFP colocalize with mCherry-TUA1 along the full lengths of the microtubules (Figure 7E). These results indicate that OsFH3 might anchor the AF cytoskeleton to microtubules, and again, that the two domains play discrete roles in microtubule binding and organization.
To understand the function of OsFH3 in changing the patterning of microtubules in rice, the microtubule staining assay was conducted. It, however, did not reveal any obvious changes of microtubule between osfh3 and WT. Notably, the osfh3 osfh5 double mutant showed similar defects of microtubule to the osfh5 single mutant, suggesting that OsFH3 does not have a key role in modulating microtubule dynamics, which differs from that of OsFH5.
References
- Suarez, C.; Carroll, R.T.; Burke, T.A.; Christensen, J.R.; Bestul, A.J.; Sees, J.A.; James, M.L.; Sirotkin, V.; Kovar, D.R. Profilin regulates F-actin network homeostasis by favoring formin over Arp2/3 complex. Dev. Cell 2015, 32, 43–53.
- Lennarz, W.J.; Lane, M.D. Encyclopedia of Biological Chemistry; Academic Press: Cambridge, MA, USA, 2013.
- Zhang, Z.; Zhang, Y.; Tan, H.; Wang, Y.; Li, G.; Liang, W.; Yuan, Z.; Hu, J.; Ren, H.; Zhang, D. RICE MORPHOLOGY DETERMINANT encodes the type II formin FH5 and regulates rice morphogenesis. Plant Cell 2011, 23, 681–700.
- Qian, D.; Xiang, Y. Actin cytoskeleton as actor in upstream and downstream of calcium signaling in plant cells. Int. J. Mol. Sci. 2019, 20, 1403.
- Li, J.; Blanchoin, L.; Staiger, C.J. Signaling to actin stochastic dynamics. Annu. Rev. Plant Biol. 2015, 66, 415–440.
- Zhao, S.; Jiang, Y.; Zhao, Y.; Huang, S.; Yuan, M.; Zhao, Y.; Guo, Y. CASEIN KINASE1-LIKE PROTEIN2 regulates actin filament stability and stomatal closure via phosphorylation of actin depolymerizing factor. Plant Cell 2016, 28, 1422–1439.
- Zhao, J. Phospholipase D and phosphatidic acid in plant defence response: From protein-protein and lipid-protein interactions to hormone signalling. J. Exp. Bot. 2015, 66, 1721–1736.
- Lian, N.; Wang, X.; Jing, Y.; Lin, J. Regulation of cytoskeleton-associated protein activities: Linking cellular signals to plant cytoskeletal function. J. Integr. Plant Biol. 2021, 63, 241–250.
- Li, P.; Day, B. Battlefield cytoskeleton: Turning the tide on plant immunity. Mol. Plant Microbe Interact. 2019, 32, 25–34.
- Li, J.; Cao, L.; Staiger, C.J. Capping protein modulates actin remodeling in response to reactive oxygen species during plant innateimmunity. Plant Physiol. 2017, 173, 1125–1136.
- Breuer, D.; Nowak, J.; Ivakov, A.; Somssich, M.; Persson, S.; Nikoloski, Z. System-wide organization of actin cytoskeleton determines organelle transport in hypocotyl plant cells. Proc. Natl. Acad. Sci. USA 2017, 114, E5741–E5749.
- Liu, C.; Zhang, Y.; Ren, H. Actin polymerization mediated by AtFH5 directs the polarity establishment and vesicle trafficking for pollen germination in Arabidopsis. Mol. Plant 2018, 11, 1389–1399.
- Li, Y.; Shen, Y.; Cai, C.; Zhong, C.; Zhu, L.; Yuan, M.; Ren, H. The type II Arabidopsis formin14 interacts with microtubules and microfilaments to regulate cell division. Plant Cell 2010, 22, 2710–2726.
- Scheuring, D.; Lofke, C.; Kruger, F.; Kittelmann, M.; Eisa, A.; Hughes, L.; Smith, R.S.; Hawes, C.; Schumacher, K.; Kleine-Vehn, J. Actin-dependent vacuolar occupancy of the cell determines auxin-induced growth repression. Proc. Natl. Acad. Sci. USA 2016, 113, 452–457.
- Edwards, M.; Zwolak, A.; Schafer, D.A.; Sept, D.; Dominguez, R.; Cooper, J.A. Capping protein regulators fine-tune actin assembly dynamics. Nat. Rev. Mol. Cell Biol. 2014, 15, 677–689.
- Chereau, D.; Dominguez, R. Understanding the role of the G-actin-binding domain of Ena/VASP in actin assembly. J. Struct. Biol. 2006, 155, 195–201.
- Goley, E.D.; Welch, M.D. The ARP2/3 complex: An actin nucleator comes of age. Nat. Rev. Mol. Cell Biol. 2006, 7, 713–726.
- Ferron, F.O.; Rebowski, G.; Lee, S.H.; Dominguez, R.J.E.J. Structural basis for the recruitment of profilin-actin complexes during filament elongation by Ena/VASP. EMBO J. 2014, 26, 4597–4606.
- García-González, J.; Kebrlová, T.; Semerák, M.; Lacek, J.; Schwarzerová, K. Arp2/3 complex is required for auxin-driven cell expansion through regulation of auxin transporter homeostasis. Front. Plant Sci. 2020, 11, 486.
- Grunt, M.; Žárský, V.; Cvrčková, F. Roots of angiosperm formins: The evolutionary history of plant FH2 domain-containing proteins. BMC Evol. Biol. 2008, 8, 115.
- Van Gisbergen, P.A.; Bezanilla, M. Plant formins: Membrane anchors for actin polymerization. Trends Cell Biol. 2013, 23, 227–233.
- Martinière, A.; Gayral, P.; Hawes, C.; Journal, J.R.J.P. Building bridges: Formin1 of Arabidopsis forms a connection between the cell wall and the actin cytoskeleton. Plant J. 2011, 66, 354–365.
- Cvrčková, F.; Novotný, M.; Pícková, D.; Žárský, V. Formin homology 2 domains occur in multiple contexts in angiosperms. BMC Genom. 2004, 5, 44.
- Chalkia, D.; Nikolaidis, N.; Makalowski, W.; Klein, J.; Nei, M.J.M.B. Evolution, origins and evolution of the formin multigene family that is involved in the formation of actin filaments. Mol. Biol. Evol. 2008, 25, 2717–2733.
- Paul, A.S.; Pollard, T.D. The role of the FH1 domain and profilin in formin-mediated actin-filament elongation and nucleation. Curr. Biol. 2008, 18, 9–19.
- Cao, L.; Henty-Ridilla, J.L.; Blanchoin, L.; Staiger, C.J. Profilin-dependent nucleation and assembly of actin filaments controls cell elongation in Arabidopsis. Plant Physiol. 2016, 170, 220–233.
- Ye, J.; Zheng, Y.; Yan, A.; Chen, N.; Wang, Z.; Huang, S.; Yang, Z. Arabidopsis formin3 directs the formation of actin cables and polarized growth in pollen tubes. Plant Cell 2009, 21, 3868–3884.
- Ingouff, M.; Gerald, J.N.F.; Guerin, C.; Robert, H.; Sorensen, M.B.; Van Damme, D.; Geelen, D.; Blanchoin, L.; Berger, F. Plant formin AtFH5 is an evolutionarily conserved actin nucleator involved in cytokinesis. Nat. Cell Biol. 2005, 7, 374–380.
- Van Gisbergen, P.; Wu, S.Z.; Cheng, X.; Pattavina, K.A.; Bezanilla, M. In vivo analysis of formin dynamics in the moss P. patens reveals functional class diversification. J. Cell Sci. 2020, 133, jcs233791.
- Oulehlová, D.; Kollárová, E.; Cifrová, P.; Pejchar, P.; Žárský, V.; Cvrčková, F. Arabidopsis class I formin FH1 relocates between membrane compartments during root cell ontogeny and associates with plasmodesmata. Plant Cell Physiol. 2019, 60, 1855–1870.
- Xue, X.; Guo, C.; Du, F.; Lu, Q.; Zhang, C.; Ren, H. AtFH8 is involved in root development under effect of low-dose latrunculin B in dividing cells. Mol. Plant 2011, 4, 264–278.
- Yi, K.; Guo, C.; Chen, D.; Zhao, B.; Yang, B.; Ren, H. Cloning and functional characterization of a formin-like protein (AtFH8) from Arabidopsis1. Plant Physiol. 2005, 138, 00001071–00001082.
- Sun, T.; Li, S.; Ren, H. OsFH15, a class I formin, interacts with microfilaments and microtubules to regulate grain size via affecting cell expansion in rice. Sci. Rep. 2017, 7, 6538.
- Cvrčková, F.; Grunt, M.; Žárský, V. Expression of GFP-mTalin reveals an actin-related role for the Arabidopsis Class II formin AtFH12. Biol. Plant. 2012, 56, 431–440.
- Vidali, L.; van Gisbergen, P.A.; Guerin, C.; Franco, P.; Li, M.; Burkart, G.M.; Augustine, R.C.; Blanchoin, L.; Bezanilla, M. Rapid formin-mediated actin-filament elongation is essential for polarized plant cell growth. Proc. Natl. Acad. Sci. USA 2009, 106, 13341–13346.
- Du, P.; Wang, J.; He, Y.; Zhang, S.; Hu, B.; Xue, X.; Miao, L.; Ren, H. AtFH14 crosslinks actin filaments and microtubules in different manners. Biol. Cell 2021, 113, 235–249.
- Kollárová, E.; Forero, A.B.; Stillerová, L.; Perostová, S.; Cvrčková, F. Arabidopsis class II formins AtFH13 and AtFH14 can form heterodimers but exhibit distinct patterns of cellular localization. Int. J. Mol. Sci 2020, 21, 348.
- Yang, W.; Ren, S.; Zhang, X.; Gao, M.; Ye, S.; Qi, Y.; Zheng, Y.; Wang, J.; Zeng, L.; Li, Q.; et al. BENT UPPERMOST INTERNODE1 encodes the class II formin FH5 crucial for actin organization and rice development. Plant Cell 2011, 23, 661–680.
- Li, G.; Liang, W.; Zhang, X.; Ren, H.; Zhang, D. Rice actin-binding protein RMD is a key link in the auxin-actin regulatory loop that controls cell growth. Proc. Natl. Acad. Sci. USA 2014, 111, 10377–10382.
- Huang, G.; Liang, W.; Sturrock, C.J.; Pandey, B.K.; Giri, J.; Mairhofer, S.; Wang, D.; Muller, L.; Tan, H.; York, L.M.; et al. Rice actin binding protein RMD controls crown root angle in response to external phosphate. Nat. Commun. 2018, 9, 2346.
- Song, Y.; Li, G.; Nowak, J.; Zhang, X.; Xu, D.; Yang, X.; Huang, G.; Liang, W.; Yang, L.; Wang, C.; et al. The rice actin-binding protein RMD regulates light-dependent shoot gravitropism. Plant Physiol. 2019, 181, 630–644.
- Kollárová, E.; Baquero Forero, A.; Cvrčková, F. The arabidopsis thaliana class II formin FH13 modulates pollen tube growth. Front. Plant Sci. 2021, 12, 599961.
- Diao, M.; Ren, S.L.; Wang, Q.N.; Qian, L.C.; Shen, J.F.; Liu, Y.L.; Huang, S.J. Arabidopsis formin 2 regulates cell-to-cell trafficking by capping and stabilizing actin filaments at plasmodesmata. Elife 2018, 7, e36316.
- Deeks, M.J.; Fendrych, M.; Smertenko, A.; Bell, K.S.; Oparka, K.; Cvrčková, F.; Žárský, V.; Hussey, P. The plant formin AtFH4 interacts with both actin and microtubules, and contains a newly identified microtubule-binding domain. J. Cell Sci. 2010, 123, 1209–1215.
More
